6 Digital soil mapping regression model
We will explore a full workflow to predict values from soil properties based on covariates.
6.1 SCORPAN model and Digital soil mapping (DSM)
 Elements:
Elements:
- S : Soil at a specific point in space and time: soil classes ( Sc ) or soil attributes ( Sa ).
From Jenny’s Equation:
- c : Climate properties of the environment.
- o : Organisms, vegetation, biotic factor.
- r : Topography, landscape attributes.
- p : Parent material, lithology.
- a : Age or time factor.
Additions:
- s : Prior knowledge of soil at a point.
- n : Prior knowledge of soil at a point.
- ℇ : Prior knowledge of soil at a point. \(\\[0.15cm]\)
- \(f()\) : Qualitative function \(f\) linking S to scorpan factors (McBratney, Mendonça Santos, and Minasny 2003).
\(\\[0.25cm]\)
At each soil sample locations the covariates cell will be observed and their value will be retain for the model training.
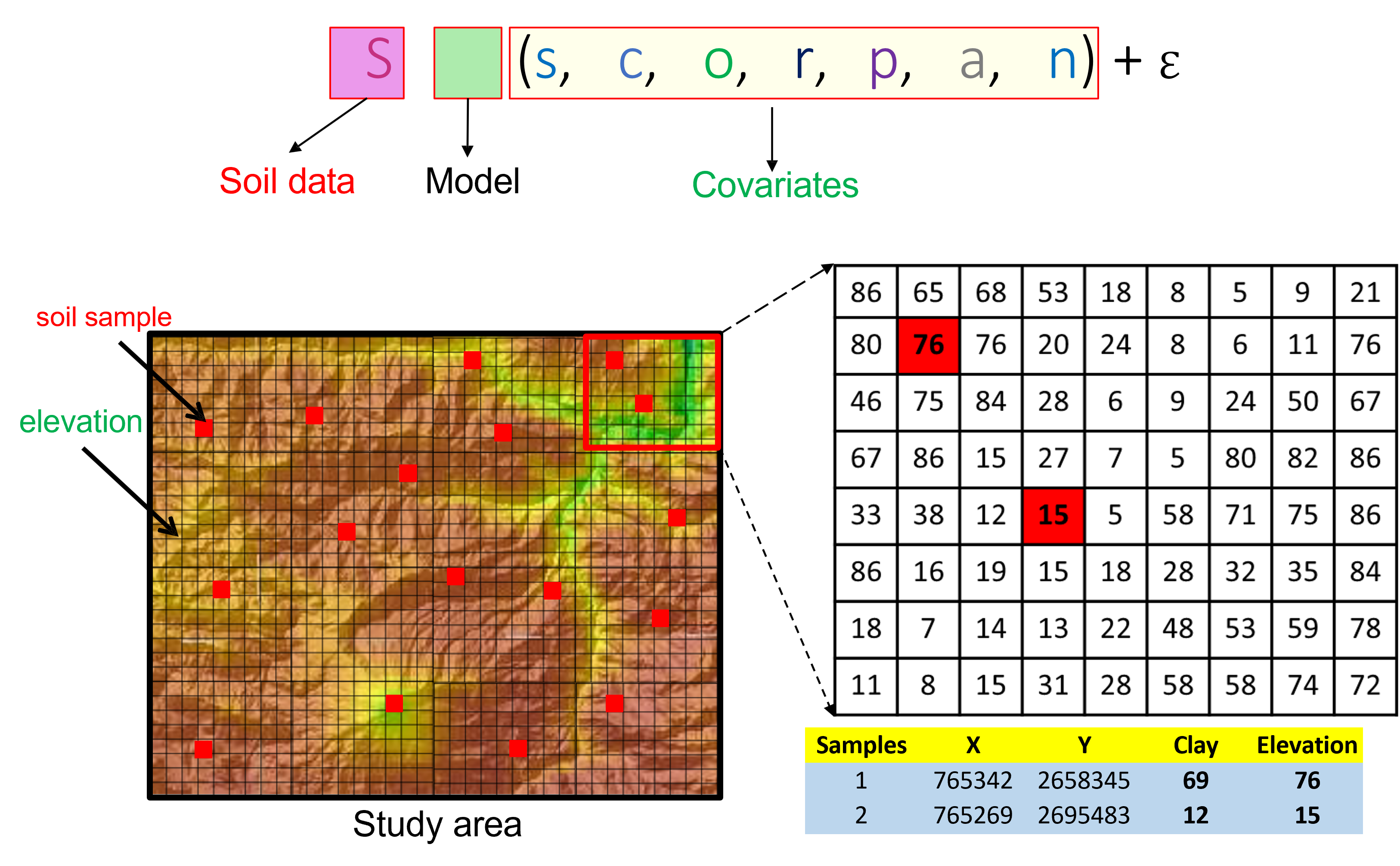
Figure 6.1: SCORPAN and relation with digital mapping.
\(\\[0.25cm]\)
Once we have several observation of the covariates a trend can be observed between variable values an samples boservation. It can follow a positive correlation or not.
\(\\[0.25cm]\)
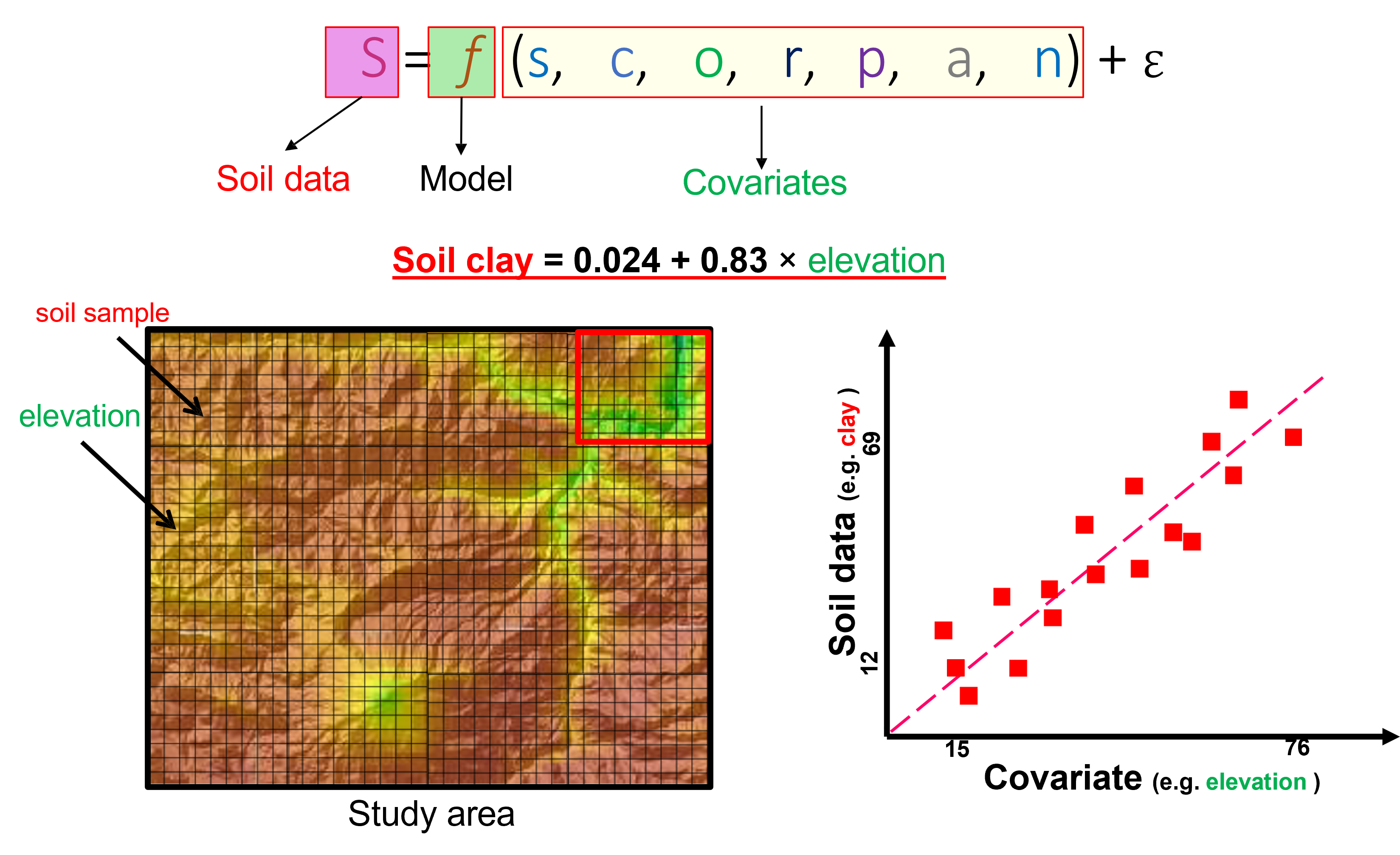
Figure 6.2: Covariates relation with soil observations.
6.2 Importing Covariates
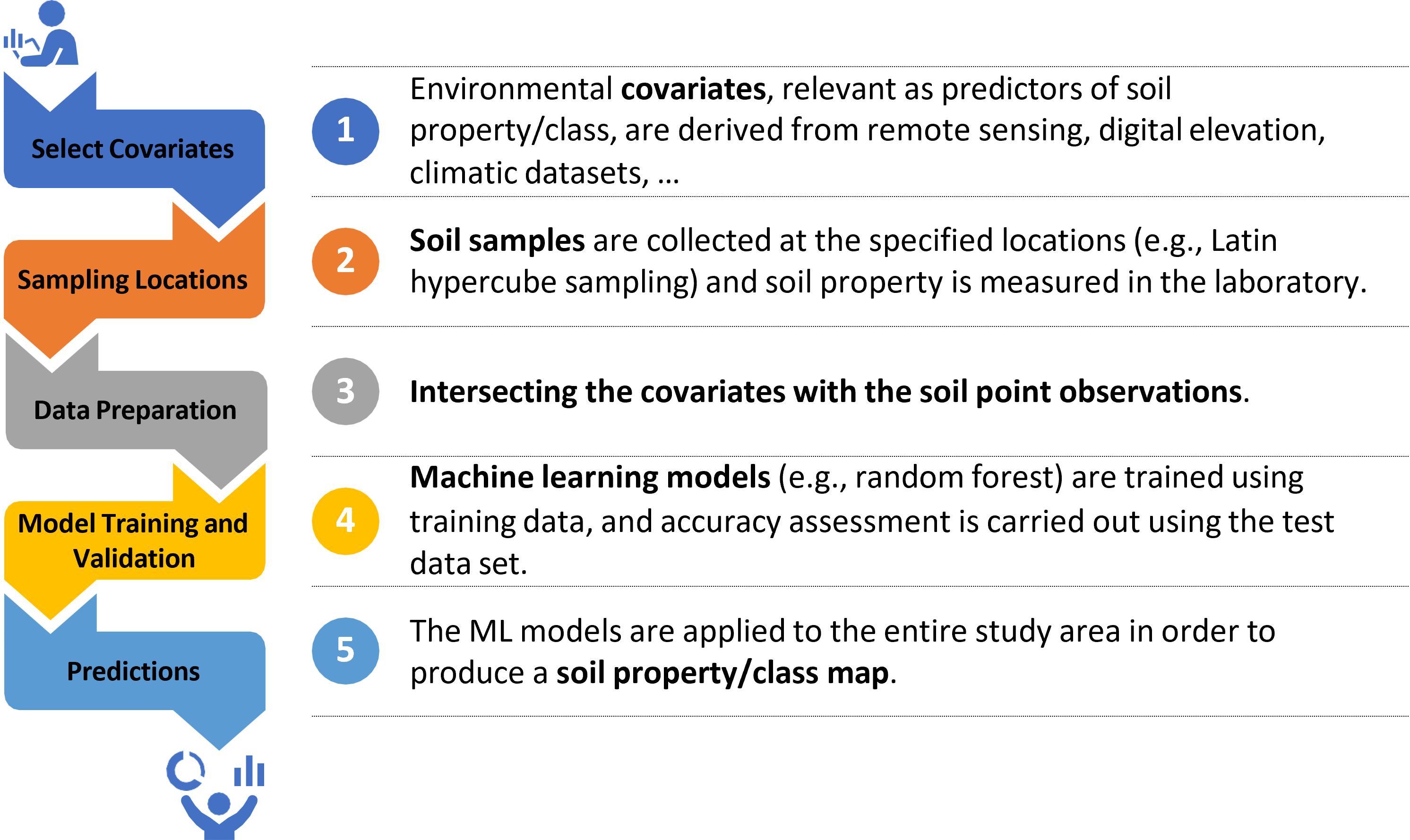
Figure 6.3: Digital soil mapping steps.
## [1] "G:/OneDrive/Ecole/Cours/GEO-77/R_Introduction/Report"## Warning: le package 'pacman' a été compilé avec la version R 4.4.1# Import covariates (Remote Sensing, Terrain, Rainfall, NPP)
# import raster layers of covariates (Remote Sensing data)
blue = raster("./Data/DSM/RS/Blue.tif")
green = raster("./Data/DSM/RS/Green.tif")
red = raster("./Data/DSM/RS/Red.tif")
nir = raster("./Data/DSM/RS/Nir.tif")6.2.1 Plot the Landsat layers
# calculate some Remote Sensing indices such as NDVI
ndvi = (nir - red) / (nir + red)
# make a stack layer from Remote Sensing data
landsat = stack(blue,green,red,nir,ndvi)
# set the names to the rasters in the stack file
names(landsat) <- c("blue","green","red","nir","ndvi")
# inspect the structure of stack layer
landsat## class : RasterStack
## dimensions : 733, 716, 524828, 5 (nrow, ncol, ncell, nlayers)
## resolution : 500, 500 (x, y)
## extent : 498000, 856000, 5235500, 5602000 (xmin, xmax, ymin, ymax)
## crs : +proj=utm +zone=32 +datum=WGS84 +units=m +no_defs
## names : blue, green, red, nir, ndvi
## min values : 47.5000000, 92.0000000, 19.5000000, 6.0000000, -0.6666667
## max values : 9.383500e+03, 1.003950e+04, 1.027300e+04, 8.635500e+03, 8.870056e-01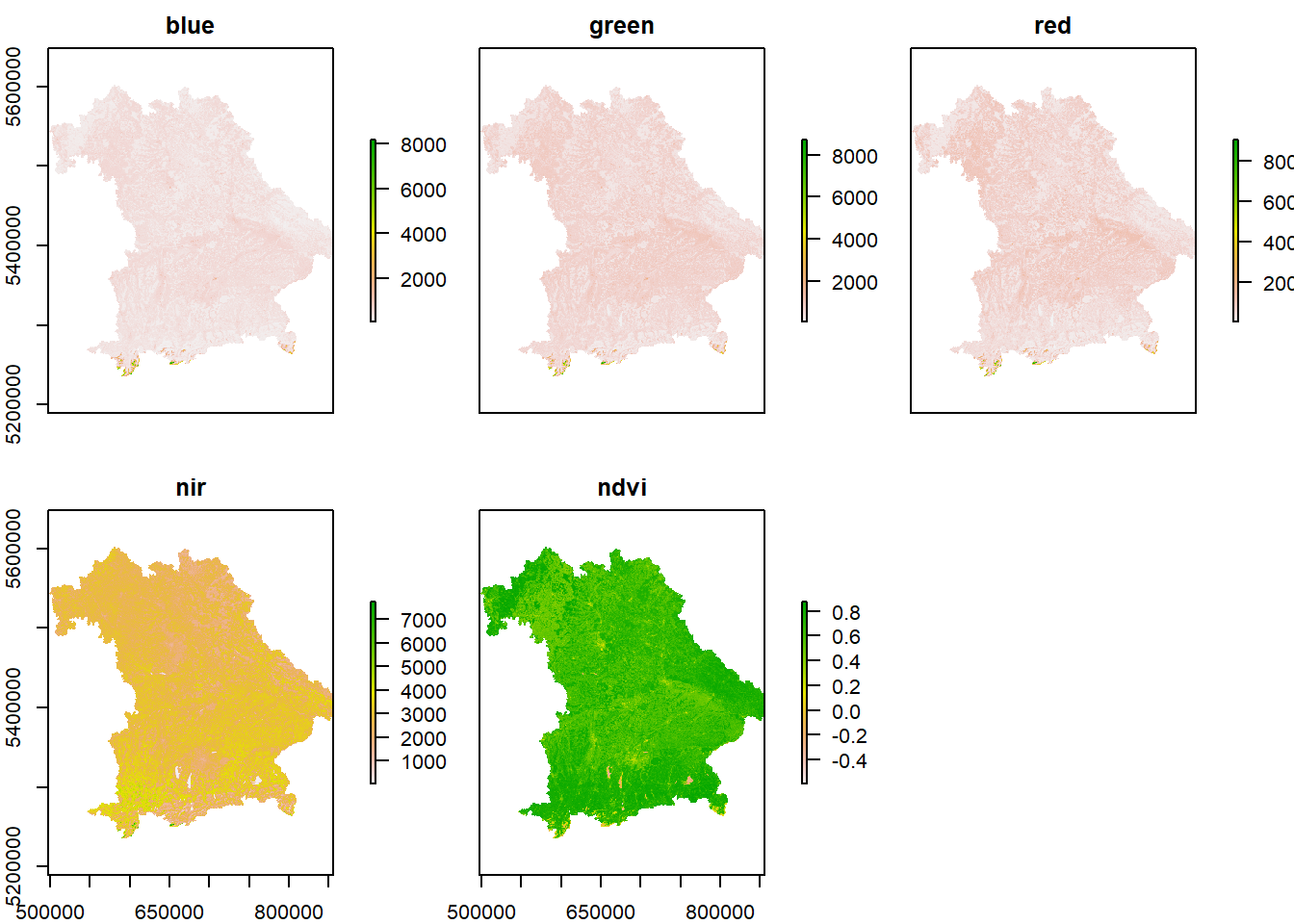
6.2.2 Stack and plot terrain variables
# import the terrain data using list.files command
dem_lst <- list.files("./Data/DSM/Terrain/", pattern="\\.sdat$", full.names = TRUE)
# inspect the structure of list
dem_lst## [1] "./Data/DSM/Terrain/CNBL.sdat" "./Data/DSM/Terrain/DEM.sdat"
## [3] "./Data/DSM/Terrain/MCA.sdat" "./Data/DSM/Terrain/Slope.sdat"
## [5] "./Data/DSM/Terrain/TWI.sdat"# make a stack layer from terrain data
terrain <- stack(dem_lst)
# plot the stack layers of terrain data
plot(terrain)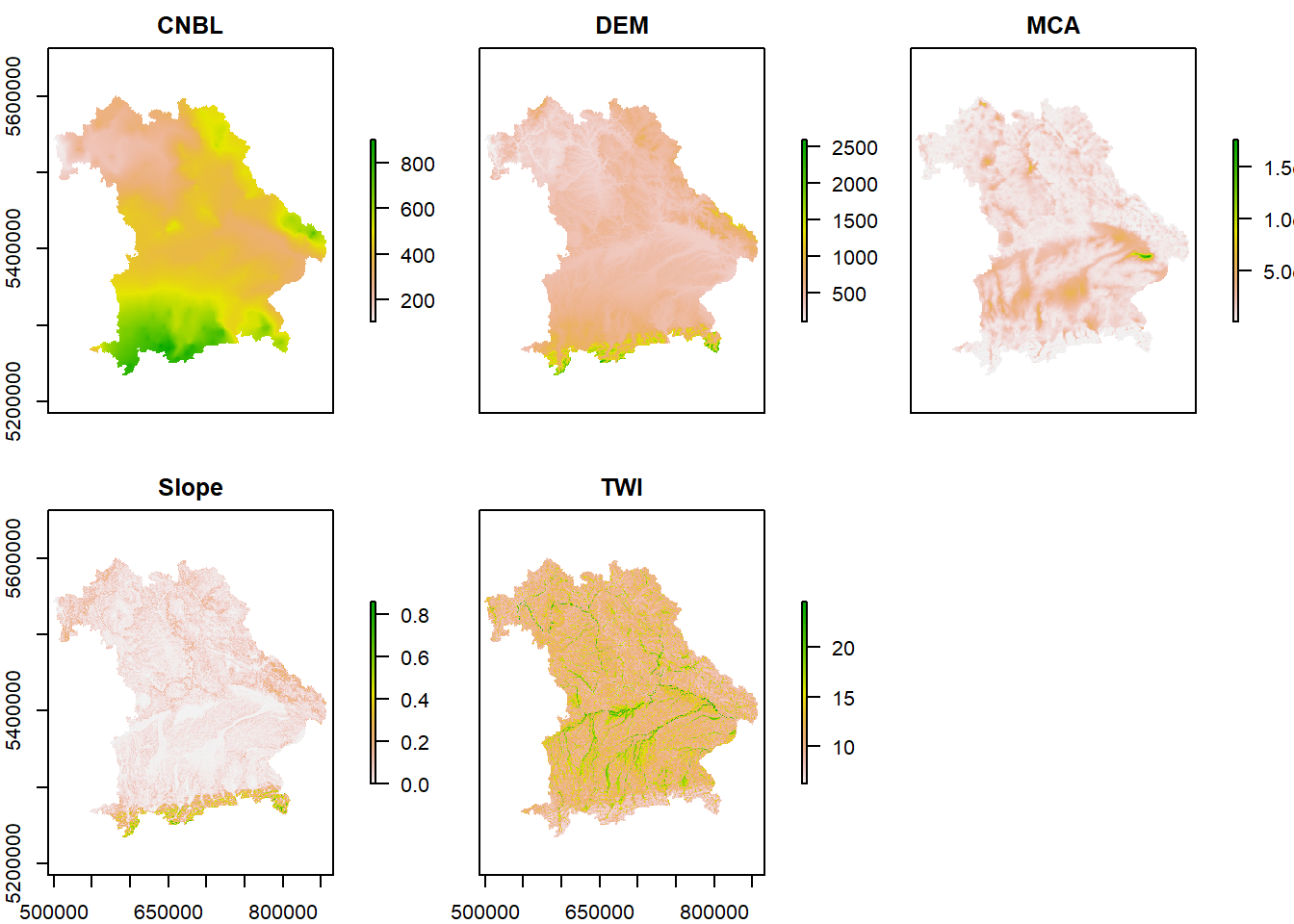
6.2.3 Plot and resample all covariates
# Re-sampling Remote Sensing data based on terrain data
RS_terrain = resample(landsat, terrain, method="bilinear")
# import mean annual rainfall (mm)
rain <- raster("./Data/DSM/Climate/rainfall.sdat")
# Re-sampling rainfall data based on terrain data
rain_terrain = resample(rain, terrain, method="bilinear")
# import Terra Net Primary Production kg*C/m^2
NPP <- raster("./Data/DSM/NPP/NPP.sdat")
# Re-sampling NPP based on terrain data
NPP_terrain = resample(NPP,terrain, method="bilinear")
# make a stack file from re-sampled remote sensing data, terrain data, rainfall, and NPP
covariates_stack = stack(terrain, RS_terrain, rain_terrain, NPP_terrain)
# inspect the structure of stack layer
names(covariates_stack)## [1] "CNBL" "DEM" "MCA" "Slope" "TWI" "blue"
## [7] "green" "red" "nir" "ndvi" "rainfall" "NPP"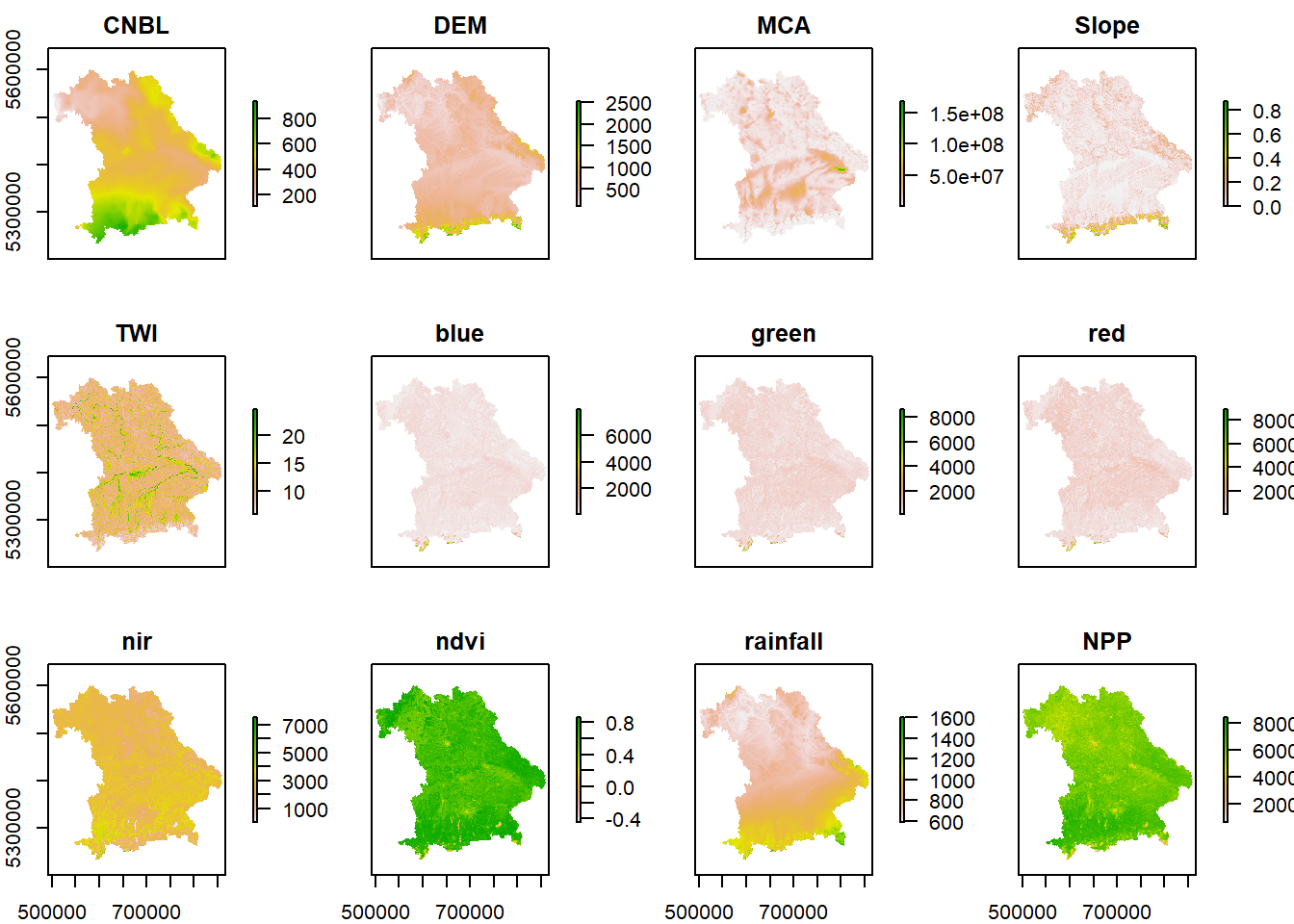
6.3 Importing study area and sampling locations
# import shape file of point data
point <- shapefile("./Data/DSM/Shapefile/points.shp")
# import shape file of Bavaria boundary
Bavaria = shapefile("./Data/DSM/Shapefile/Bavaria.shp")
# plot the point on the raster
plot(covariates_stack$DEM, main = "DEM", xlab = "Easting (m)", ylab = "Northing (m)")
plot(point,add =T, pch = 19)
plot(Bavaria, add =T)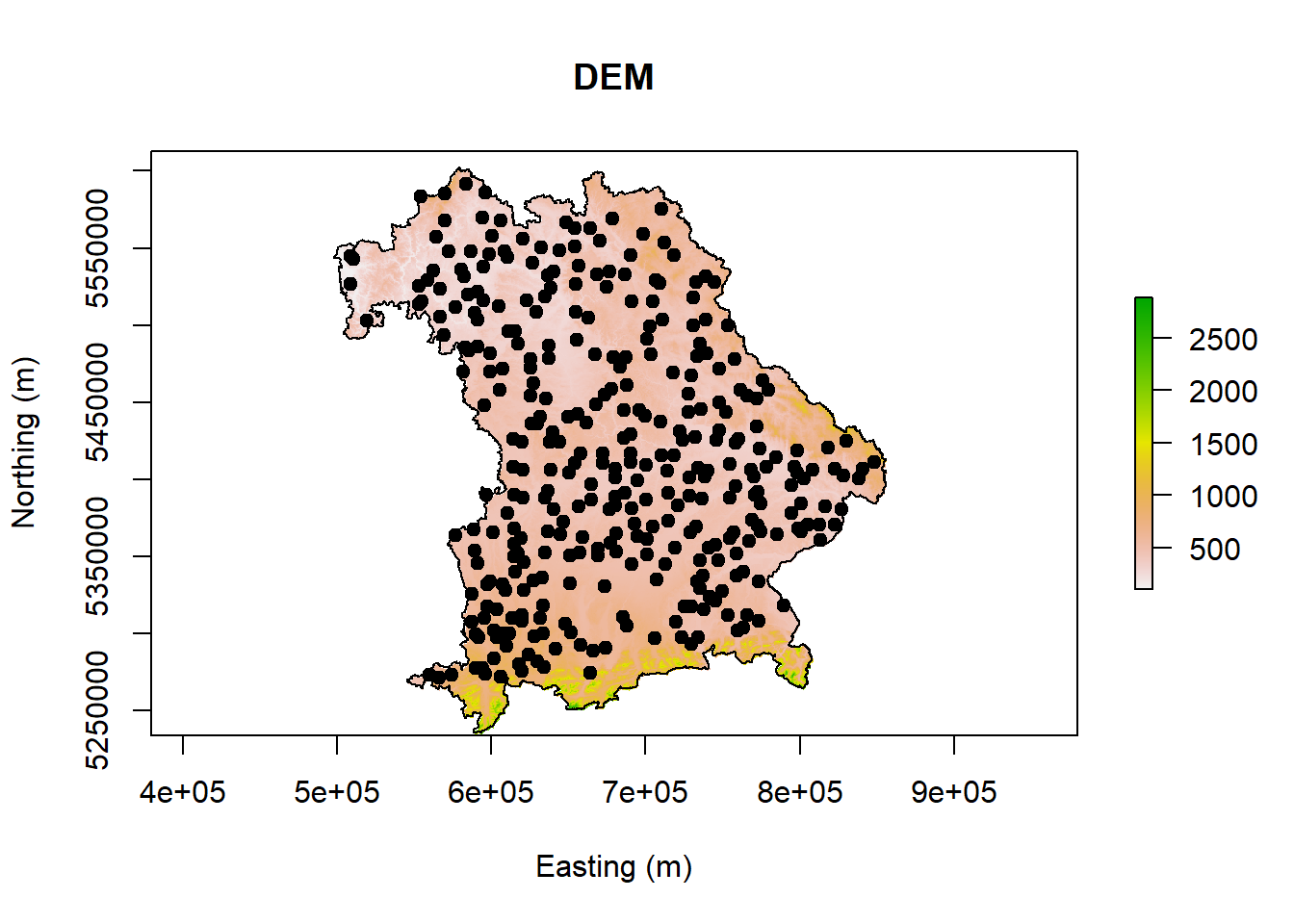
# plot an interactive map
mapview::mapview(point, zcol = "OC", at = c(0,5,10,15,20,25,30,35,200), legend = TRUE)\(\\[0.15cm]\)
## [1] "SpatialPointsDataFrame"
## attr(,"package")
## [1] "sp"## Object of class SpatialPointsDataFrame
## Coordinates:
## min max
## x 508615.3 848381.1
## y 5271503.2 5591741.8
## Is projected: TRUE
## proj4string :
## [+proj=utm +zone=32 +datum=WGS84 +units=m +no_defs]
## Number of points: 346
## Data attributes:
## x y pH OC
## Min. :508615 Min. :5271503 Min. :3.810 Min. : 6.00
## 1st Qu.:617164 1st Qu.:5352240 1st Qu.:6.162 1st Qu.: 14.50
## Median :672285 Median :5406629 Median :6.850 Median : 21.00
## Mean :676277 Mean :5416121 Mean :6.738 Mean : 28.35
## 3rd Qu.:733674 3rd Qu.:5485267 3rd Qu.:7.380 3rd Qu.: 33.05
## Max. :848381 Max. :5591742 Max. :8.140 Max. :251.30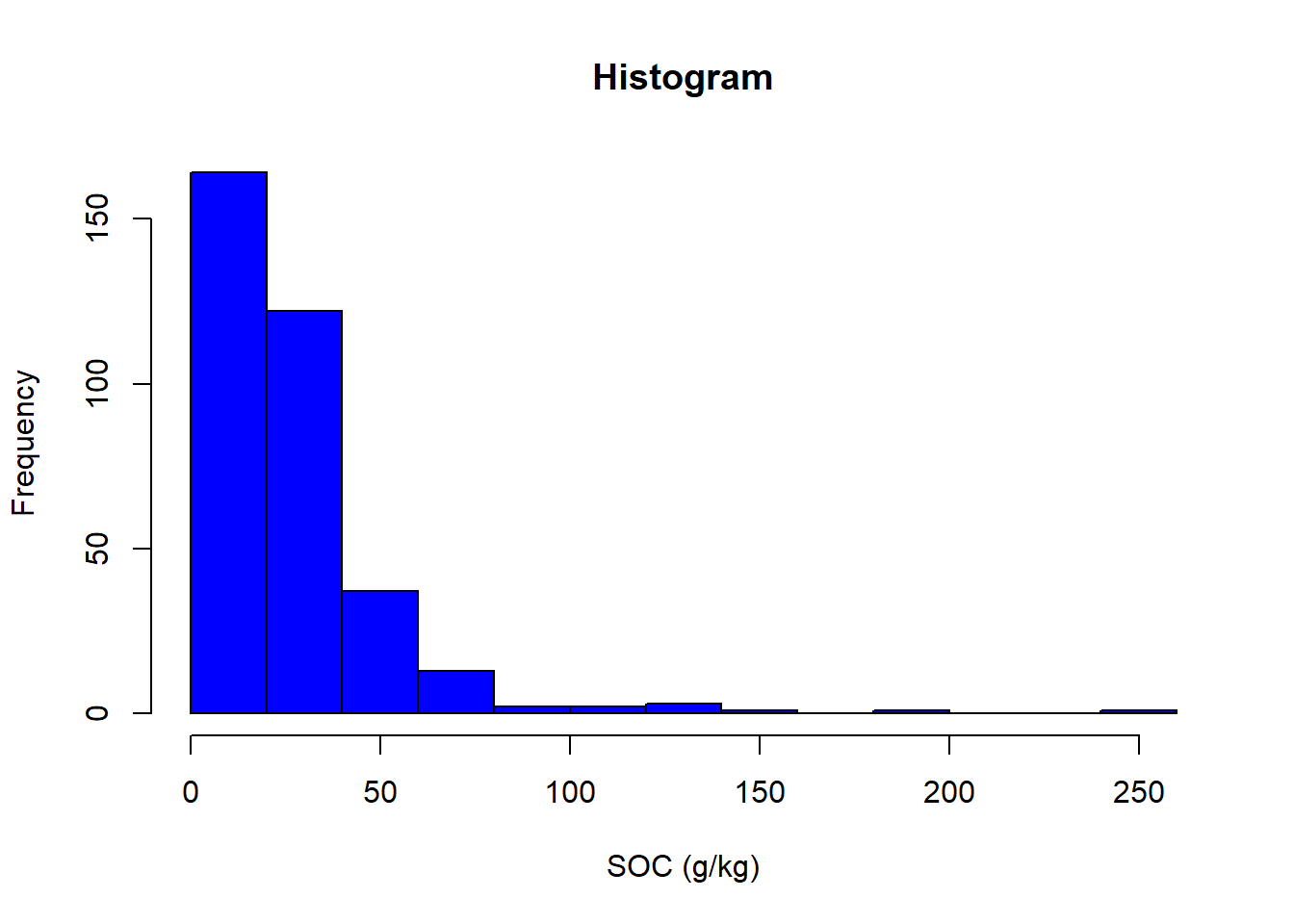
6.4 Extract the raster values at point locations
# extract covariate values at each point of observation
cov = extract(covariates_stack, point, method='bilinear', df=TRUE)
# inspect the data.frame of cov
str(cov)## 'data.frame': 346 obs. of 13 variables:
## $ ID : num 1 2 3 4 5 6 7 8 9 10 ...
## $ CNBL : num 398 431 587 408 535 ...
## $ DEM : num 426 493 600 474 544 ...
## $ MCA : num 6744760 14292214 4667933 5777121 14213043 ...
## $ Slope : num 0.0465 0.0149 0.026 0.0228 0.016 ...
## $ TWI : num 10.5 11.1 13.2 11.3 12 ...
## $ blue : num 236 401 374 375 411 ...
## $ green : num 403 698 599 670 680 ...
## $ red : num 273 680 537 586 544 ...
## $ nir : num 2561 3008 2808 3300 3390 ...
## $ ndvi : num 0.811 0.632 0.663 0.699 0.725 ...
## $ rainfall: num 1105 1039 1105 964 1051 ...
## $ NPP : num 7384 6576 6772 6682 6919 ...# combining covariates and soil properties
cov_soil = cbind(cov[,-1], OC=point$OC)
# inspect the data.frame of cov_soil
str(cov_soil)## 'data.frame': 346 obs. of 13 variables:
## $ CNBL : num 398 431 587 408 535 ...
## $ DEM : num 426 493 600 474 544 ...
## $ MCA : num 6744760 14292214 4667933 5777121 14213043 ...
## $ Slope : num 0.0465 0.0149 0.026 0.0228 0.016 ...
## $ TWI : num 10.5 11.1 13.2 11.3 12 ...
## $ blue : num 236 401 374 375 411 ...
## $ green : num 403 698 599 670 680 ...
## $ red : num 273 680 537 586 544 ...
## $ nir : num 2561 3008 2808 3300 3390 ...
## $ ndvi : num 0.811 0.632 0.663 0.699 0.725 ...
## $ rainfall: num 1105 1039 1105 964 1051 ...
## $ NPP : num 7384 6576 6772 6682 6919 ...
## $ OC : num 17.8 18 27.7 12.5 31.5 ...# check the correlation covariates and OC
corrplot.mixed(cor(cov_soil), lower.col = "black", number.cex = .7)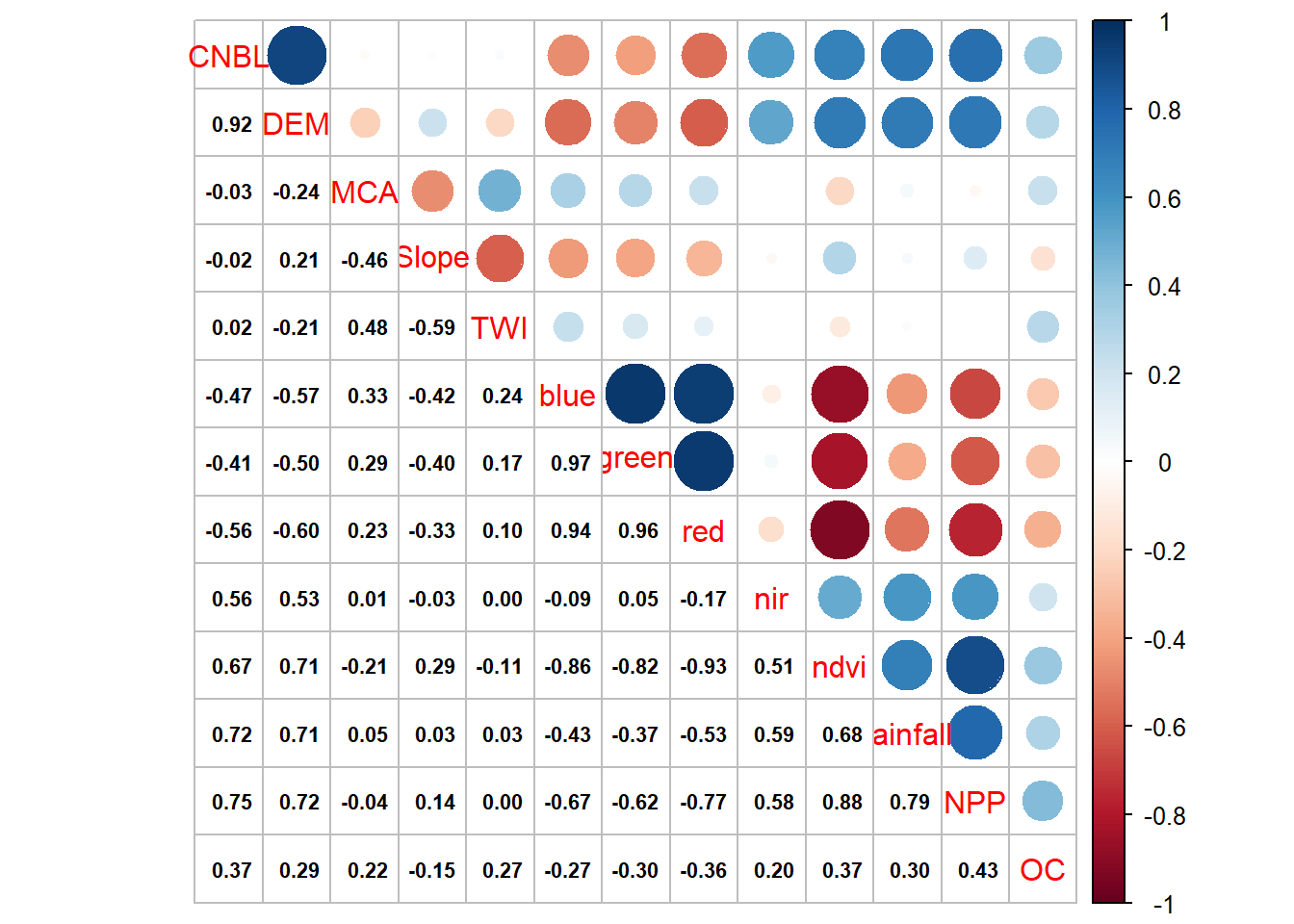
6.5 Prepare the data set before machine learning training
createDataPartition command to divide the data set into a p = split. It will create an index from which you will need to select the row belonging or not to the value.
set.seed allow you to create a specific randomness. Useful to reproduce an experiment.
# remove na values
cov_soil <- cov_soil[complete.cases(cov_soil),]
# remove high values of oc
cov_soil <- cov_soil[cov_soil$OC<78,]
# check number of rows
nrow(cov_soil)## [1] 336## [1] 13# split the data to training (80%) and testing (20%) sets
set.seed(1234)
trainIndex <- createDataPartition(cov_soil$OC,
p = 0.8, list = FALSE, times = 1)
# subset the datasets
cov_soil_Train <- cov_soil[ trainIndex,]
cov_soil_Test <- cov_soil[-trainIndex,]
# inspect the two datasets
str(cov_soil_Train)## 'data.frame': 271 obs. of 13 variables:
## $ CNBL : num 431 587 535 414 403 ...
## $ DEM : num 493 600 544 466 408 ...
## $ MCA : num 14292214 4667933 14213043 18620790 22964809 ...
## $ Slope : num 0.01492 0.02598 0.01596 0.00466 0.00203 ...
## $ TWI : num 11.1 13.2 12 13 14.1 ...
## $ blue : num 401 374 411 472 489 ...
## $ green : num 698 599 680 810 806 ...
## $ red : num 680 537 544 783 797 ...
## $ nir : num 3008 2808 3390 3573 3300 ...
## $ ndvi : num 0.632 0.663 0.725 0.64 0.611 ...
## $ rainfall: num 1039 1105 1051 984 931 ...
## $ NPP : num 6576 6772 6919 6549 6491 ...
## $ OC : num 18 27.7 31.5 12.4 17 72.8 15.2 30.7 17.4 20.2 ...## 'data.frame': 65 obs. of 13 variables:
## $ CNBL : num 398 408 420 422 485 ...
## $ DEM : num 426 474 485 502 496 ...
## $ MCA : num 6744760 5777121 21438967 7046277 11136973 ...
## $ Slope : num 0.0465 0.0228 0.0103 0.0194 0.0192 ...
## $ TWI : num 10.5 11.3 11.3 10.4 14.1 ...
## $ blue : num 236 375 421 317 353 ...
## $ green : num 403 670 742 580 613 ...
## $ red : num 273 586 690 512 482 ...
## $ nir : num 2561 3300 3420 2784 3538 ...
## $ ndvi : num 0.811 0.699 0.666 0.693 0.758 ...
## $ rainfall: num 1105 964 983 928 972 ...
## $ NPP : num 7384 6682 6859 6529 7115 ...
## $ OC : num 17.8 12.5 36.1 19.1 57.1 41.1 15 20.1 14.9 37.7 ...6.6 Train the linear regression
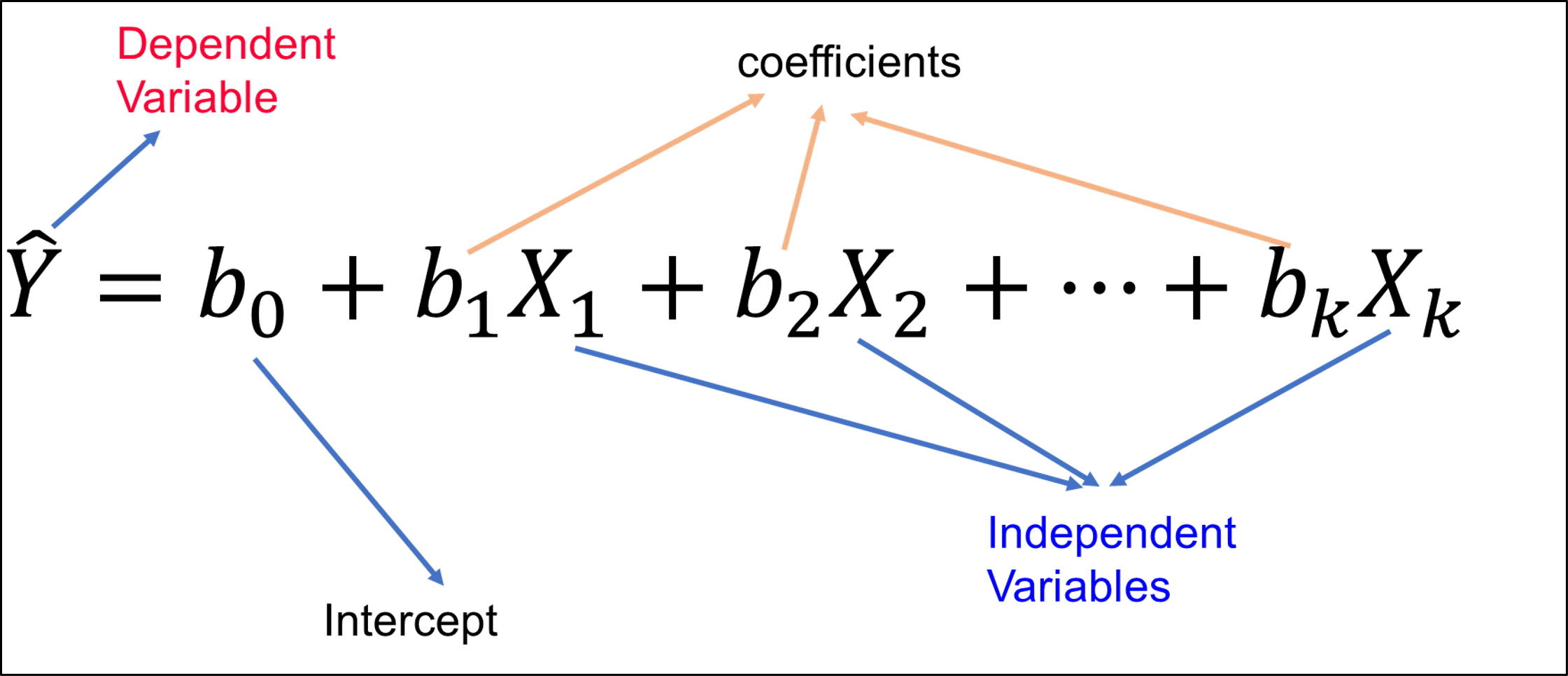
Figure 6.4: Linear regression schema.
lm(y ~ X, data = df) create a linear regression between y variables and X predictors (several columns) from the chosen data frame df.
predict(model, test) predict the test values based on the train model. They need to have the same format (ncol, and colnames) as the training data.
# fit a linear regression on training set
set.seed(1234)
linear_fit <- lm(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data=cov_soil_Train)
# look at the summary of linear model
summary(linear_fit)##
## Call:
## lm(formula = OC ~ CNBL + DEM + MCA + Slope + TWI + blue + green +
## red + nir + ndvi + rainfall + NPP, data = cov_soil_Train)
##
## Residuals:
## Min 1Q Median 3Q Max
## -22.735 -7.836 -1.557 6.239 49.203
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.693e+02 5.694e+01 2.973 0.003225 **
## CNBL -2.573e-02 2.026e-02 -1.270 0.205214
## DEM 5.678e-02 1.928e-02 2.945 0.003526 **
## MCA 2.969e-07 8.091e-08 3.670 0.000295 ***
## Slope -7.782e+01 4.139e+01 -1.880 0.061228 .
## TWI 1.317e+00 3.987e-01 3.305 0.001085 **
## blue 7.050e-02 4.455e-02 1.583 0.114753
## green -3.596e-02 5.618e-02 -0.640 0.522727
## red -1.473e-01 3.888e-02 -3.790 0.000188 ***
## nir 2.749e-02 8.490e-03 3.237 0.001365 **
## ndvi -2.306e+02 8.539e+01 -2.701 0.007369 **
## rainfall -2.693e-02 1.032e-02 -2.611 0.009567 **
## NPP 5.853e-04 3.201e-03 0.183 0.855045
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 11.51 on 258 degrees of freedom
## Multiple R-squared: 0.443, Adjusted R-squared: 0.4171
## F-statistic: 17.1 on 12 and 258 DF, p-value: < 2.2e-16# apply the linear model on testing data
OC_linear_Pred <- predict(linear_fit, cov_soil_Test)
# check the plot actual and predicted OC values
plot(cov_soil_Test$OC, OC_linear_Pred, main="Linear model",
col="blue",xlab="Actual OC", ylab="Predicted OC",
xlim=c(0,100),ylim=c(0,100))
# Create a line of 1/1 showing the "perfect" prediction rate
abline(coef = c(0,1), col="red" ) 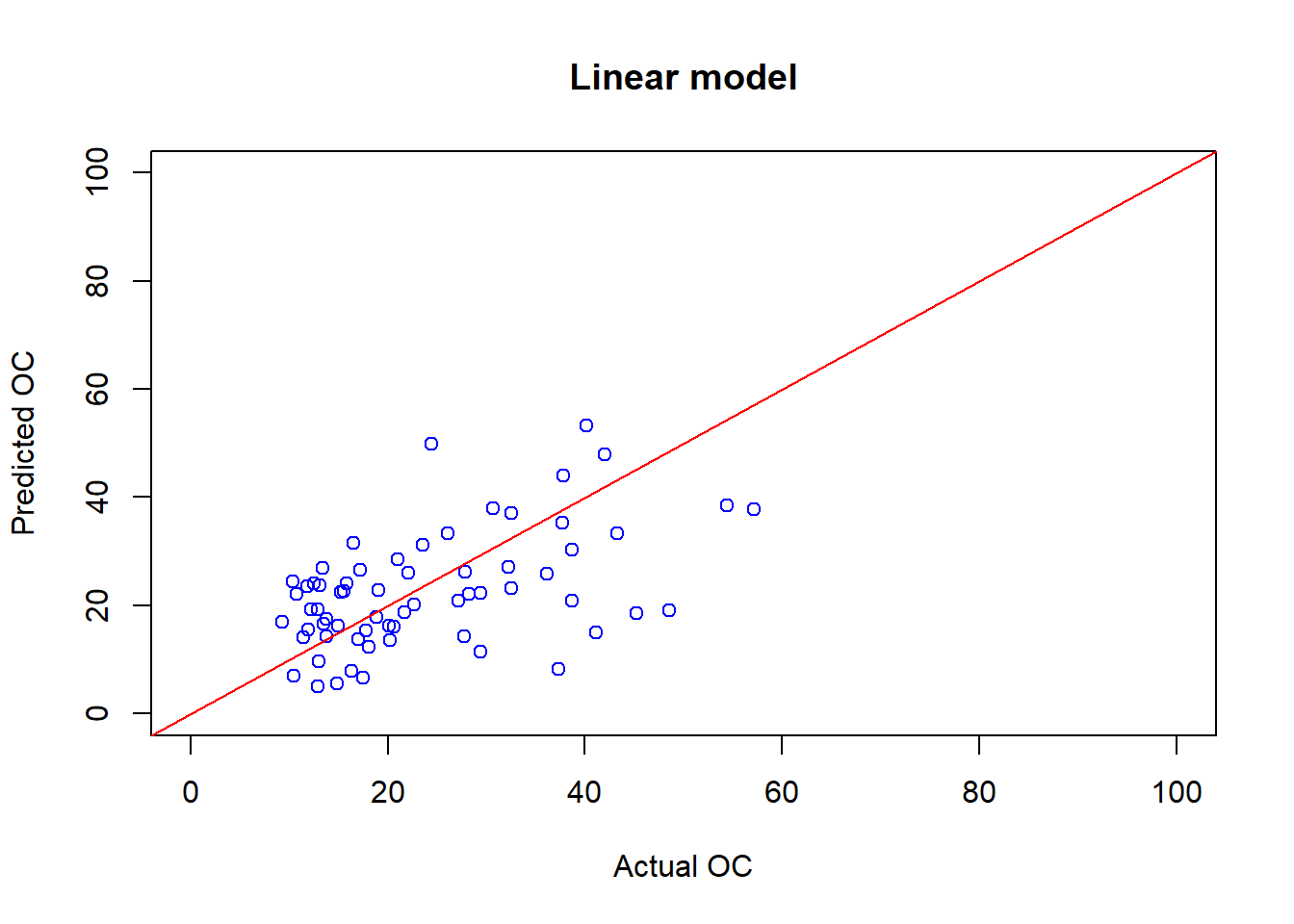
## [1] 0.4996546## [1] 11.304826.7 Train the decision tree
![Decision tree schema [@alpaydin_introduction_2014].](images/Decision_tree_model.png)
Figure 6.5: Decision tree schema (Alpaydin 2014).
rpart(y ~ X, method = "", data = df) create a decision tree for the y variables and X predictors (several columns) from the chosen data frame df. The method parameter is by default “anova” but it can be change to “poisson” in case of discrete values. For classification problems the method can be set to “class”.
# fit decision tree model on training set
set.seed(1234)
tree_fit <- rpart(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
method="anova", data=cov_soil_Train)
# display the results of decision tree
printcp(tree_fit) ##
## Regression tree:
## rpart(formula = OC ~ CNBL + DEM + MCA + Slope + TWI + blue +
## green + red + nir + ndvi + rainfall + NPP, data = cov_soil_Train,
## method = "anova")
##
## Variables actually used in tree construction:
## [1] blue CNBL DEM green MCA ndvi NPP red Slope TWI
##
## Root node error: 61351/271 = 226.39
##
## n= 271
##
## CP nsplit rel error xerror xstd
## 1 0.189128 0 1.00000 1.01112 0.104709
## 2 0.068565 1 0.81087 0.84893 0.097943
## 3 0.061621 2 0.74231 0.83187 0.096378
## 4 0.043476 3 0.68069 0.81398 0.094965
## 5 0.034078 4 0.63721 0.80606 0.095067
## 6 0.033352 5 0.60313 0.80303 0.094535
## 7 0.030887 6 0.56978 0.80303 0.094535
## 8 0.025692 7 0.53889 0.84399 0.099558
## 9 0.011202 8 0.51320 0.79923 0.091556
## 10 0.010799 10 0.49080 0.76902 0.083771
## 11 0.010465 11 0.48000 0.76902 0.083771
## 12 0.010000 12 0.46953 0.76559 0.083593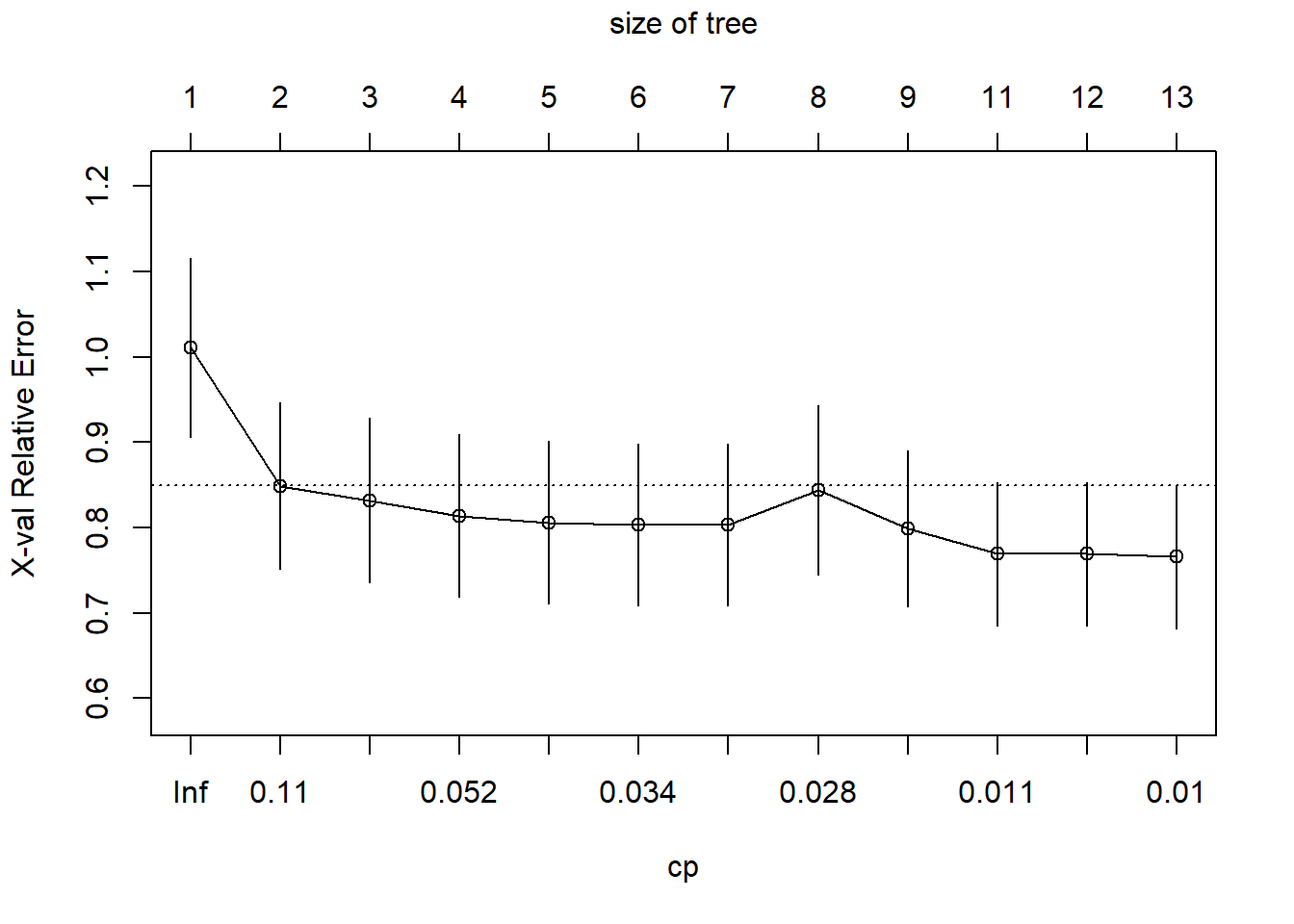
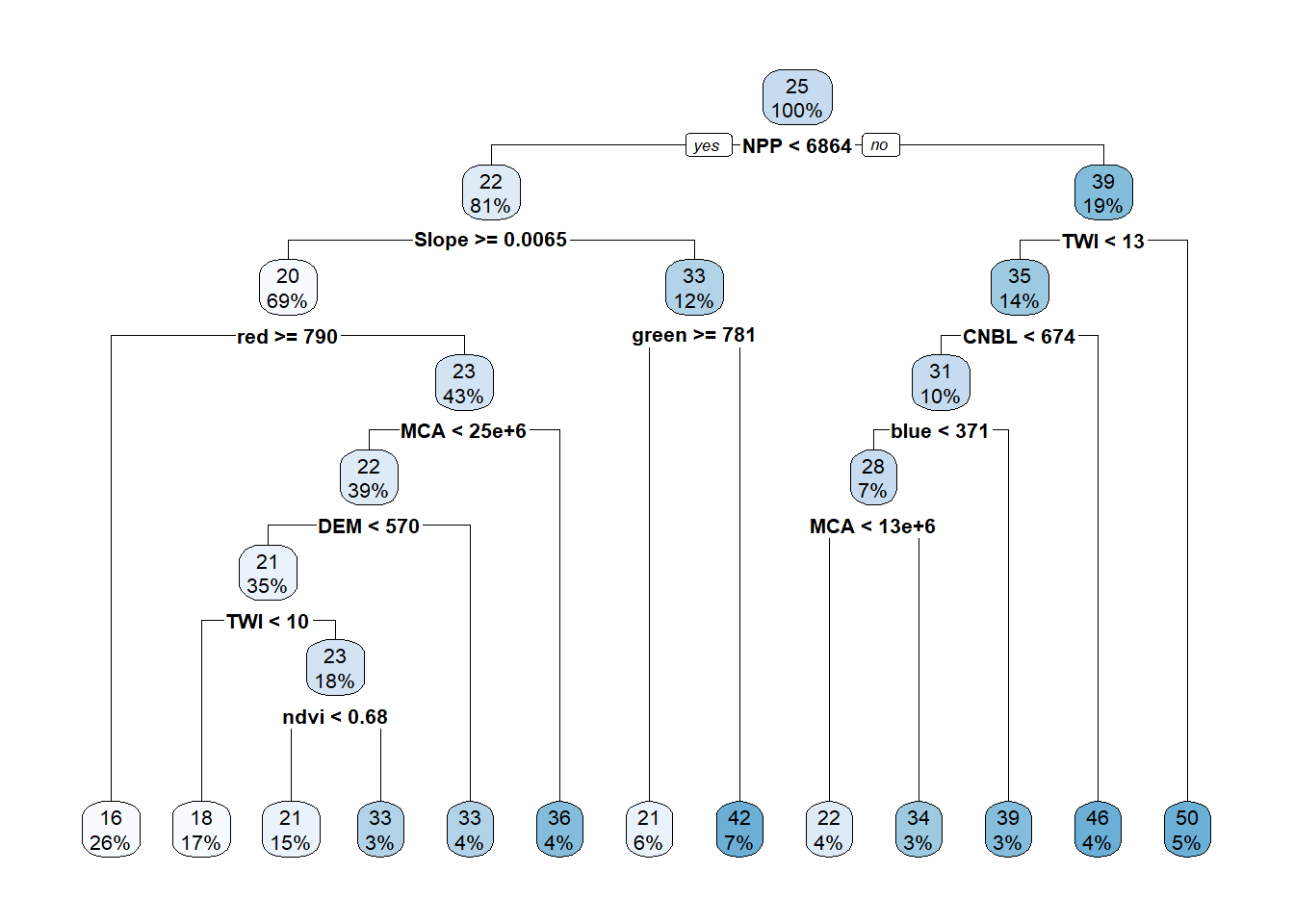
# apply the tree model on testing data
OC_tree_Pred <- predict(tree_fit, cov_soil_Test)
# check the plot actual and predicted OC values
plot(cov_soil_Test$OC, OC_tree_Pred, main="Tree model",
col="blue",xlab="Actual OC", ylab="Predicted OC", xlim=c(0,100),ylim=c(0,100))
abline(coef = c(0,1), col="red" )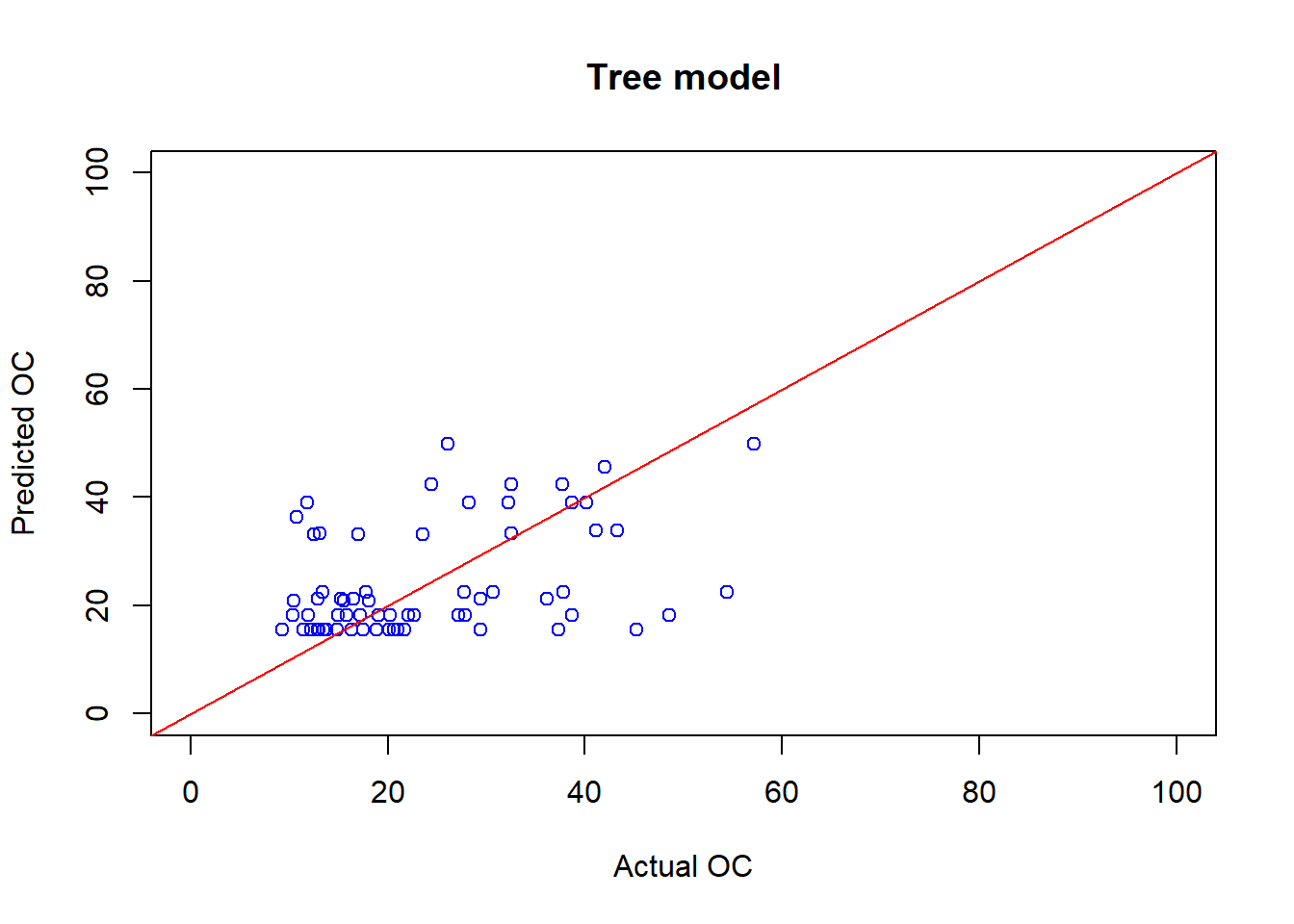
## [1] 0.4036307## [1] 12.028236.8 Train the Random forest
The random forest will produce several single decision trees and aggregate the results.
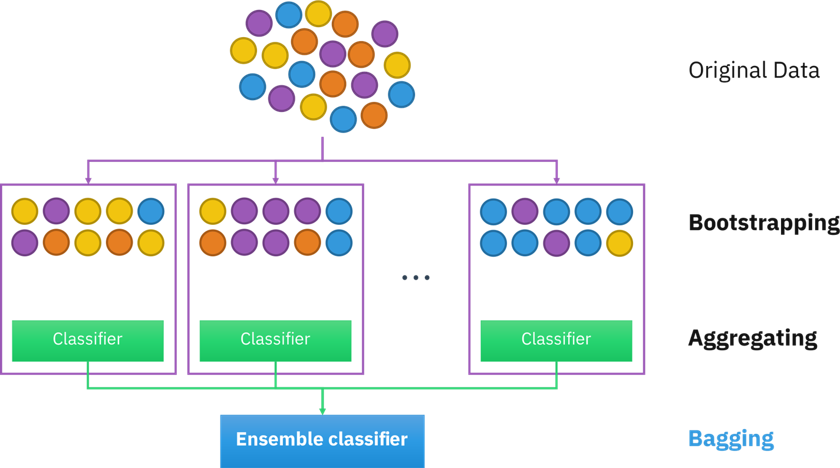
Figure 6.6: Random forest model.
randomForest(y ~ X, data = df) create a decision y variables and X predictors (several columns) from the chosen data frame df.
# fit random forest model on training set
set.seed(1234)
rf_fit <- randomForest(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data=cov_soil_Train, ntree=500, do.trace = 25)## | Out-of-bag |
## Tree | MSE %Var(y) |
## 25 | 149.4 65.98 |
## 50 | 145.5 64.25 |
## 75 | 143.2 63.27 |
## 100 | 146.4 64.68 |
## 125 | 146.4 64.67 |
## 150 | 144.8 63.97 |
## 175 | 143.9 63.58 |
## 200 | 143.4 63.34 |
## 225 | 143.3 63.29 |
## 250 | 143.6 63.44 |
## 275 | 144 63.61 |
## 300 | 144.1 63.67 |
## 325 | 143.7 63.49 |
## 350 | 143.7 63.48 |
## 375 | 143.5 63.40 |
## 400 | 143.2 63.26 |
## 425 | 143 63.17 |
## 450 | 142.9 63.14 |
## 475 | 143 63.15 |
## 500 | 142.9 63.12 |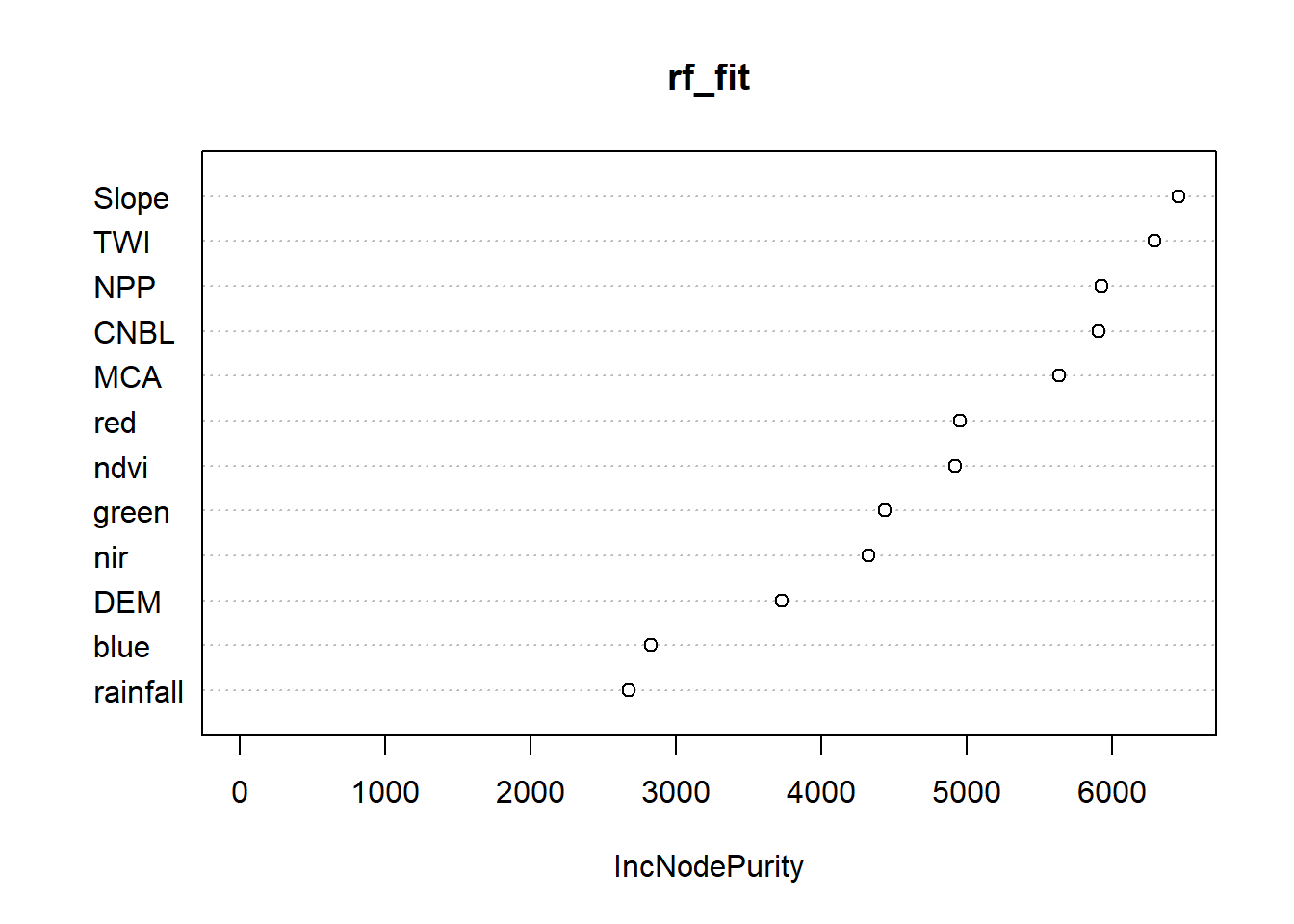
# apply the random forest model on testing data
OC_rf_Pred <- predict(rf_fit, cov_soil_Test)
# check the plot actual and predicted OC values
plot(cov_soil_Test$OC, OC_rf_Pred, main="Tree model",
col="blue",xlab="Actual OC", ylab="Predicted OC", xlim=c(0,100),ylim=c(0,100))
abline(coef = c(0,1), col="red" )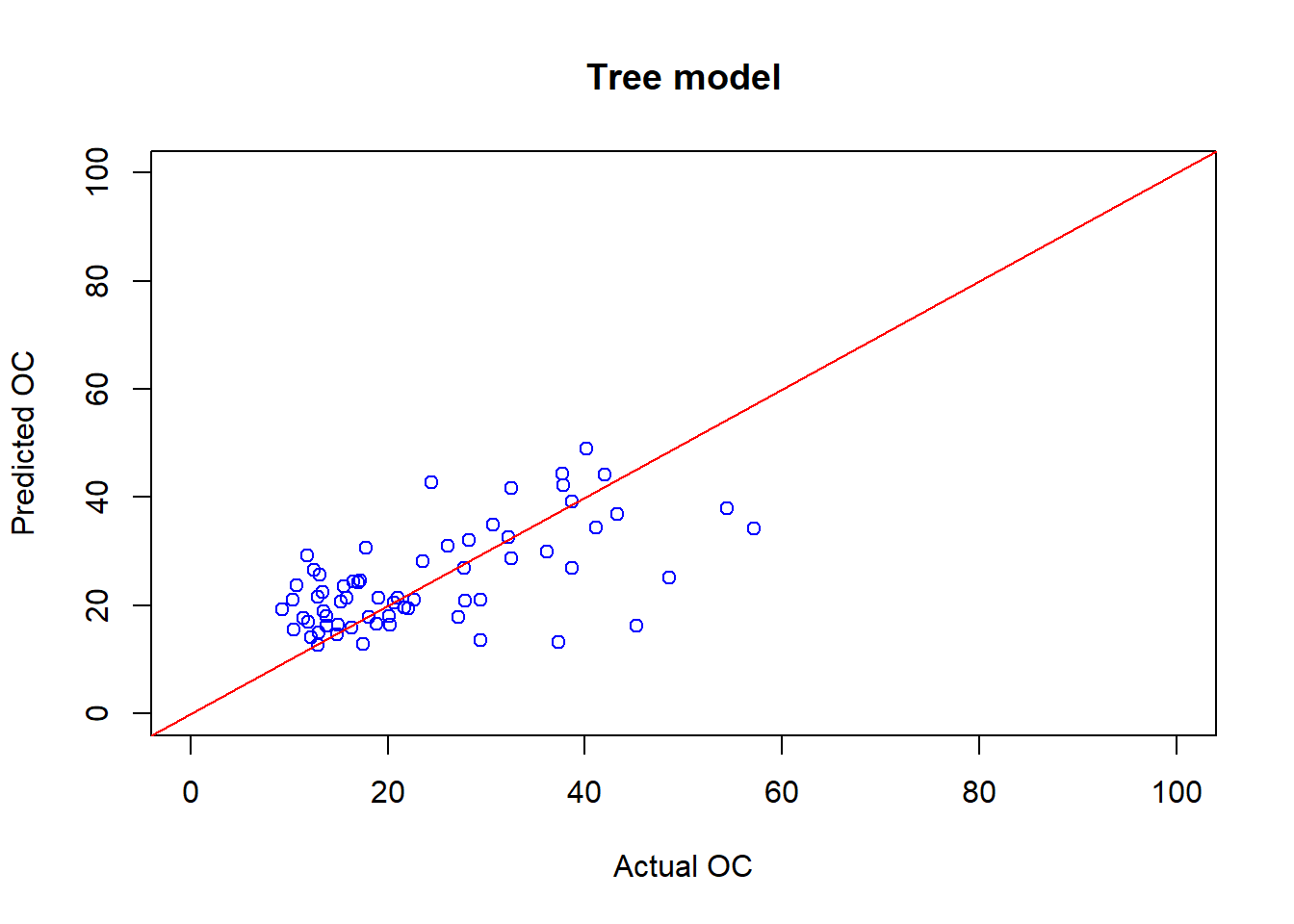
## [1] 0.6036493## [1] 9.6171226.9 Comparing the results
We have to compare the different results of the models with RMSE and R² values.
# check the accuracy of three models
RMSE_models <- c(Linear=RMSE_linear,Tree=RMSE_tree,RF=RMSE_rf)
cor_models <- c(Linear=cor_linear,Tree=cor_tree,RF=cor_rf)
# plot the final results
par(mfrow=c(1,2))
barplot(RMSE_models, main="RMSE",col=c("red","blue","green"))
barplot(cor_models, main="Correlation",col=c("red","blue","green"))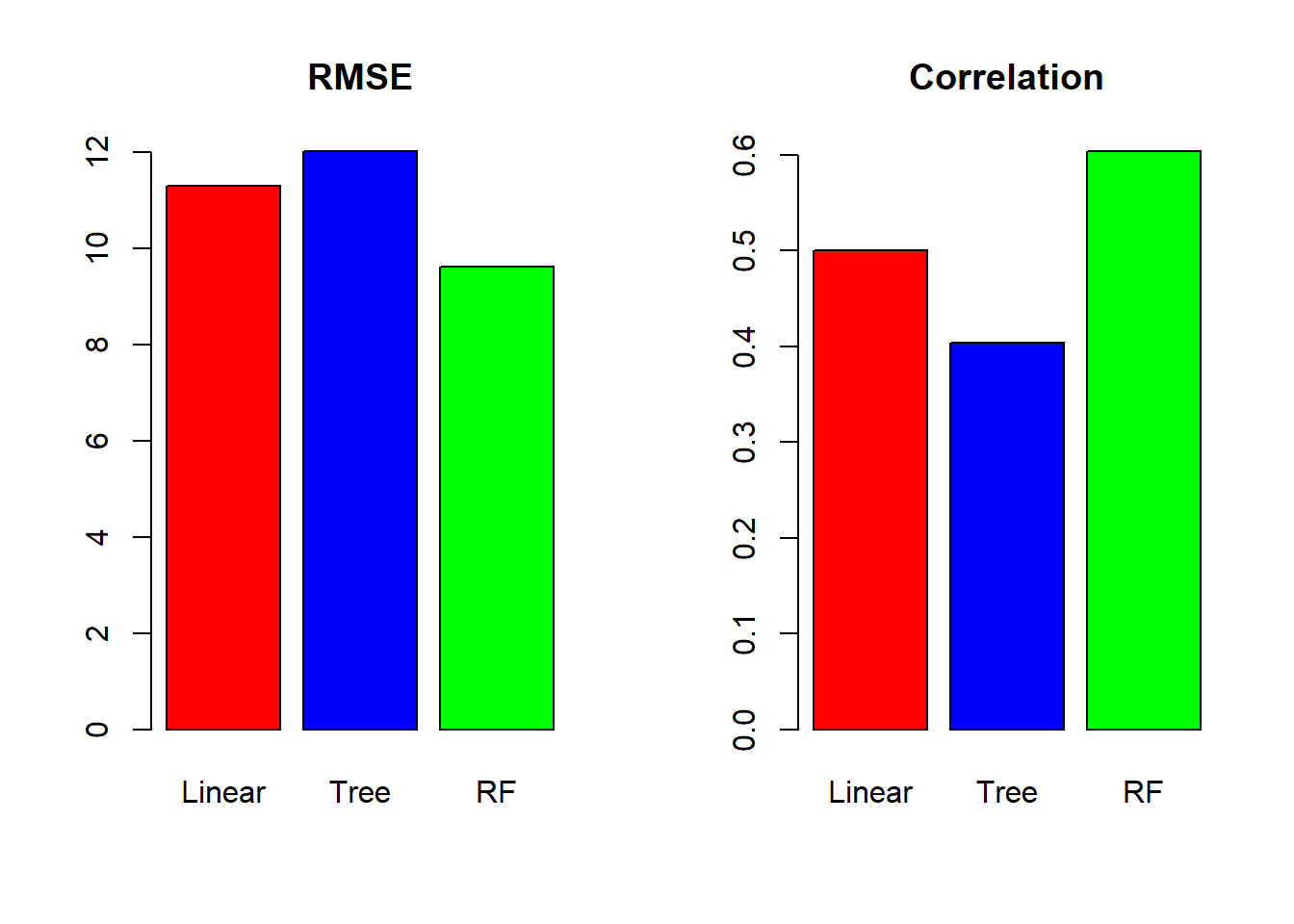
6.10 Create the prediction map
We are choosing the model with the best performances (RF). When predicting you do not need to set a seed.
# apply the best model on the stack layer
map_rf <- raster::predict(covariates_stack, rf_fit)
# plot the final map
spplot(map_rf, main="SOC map based on RF model")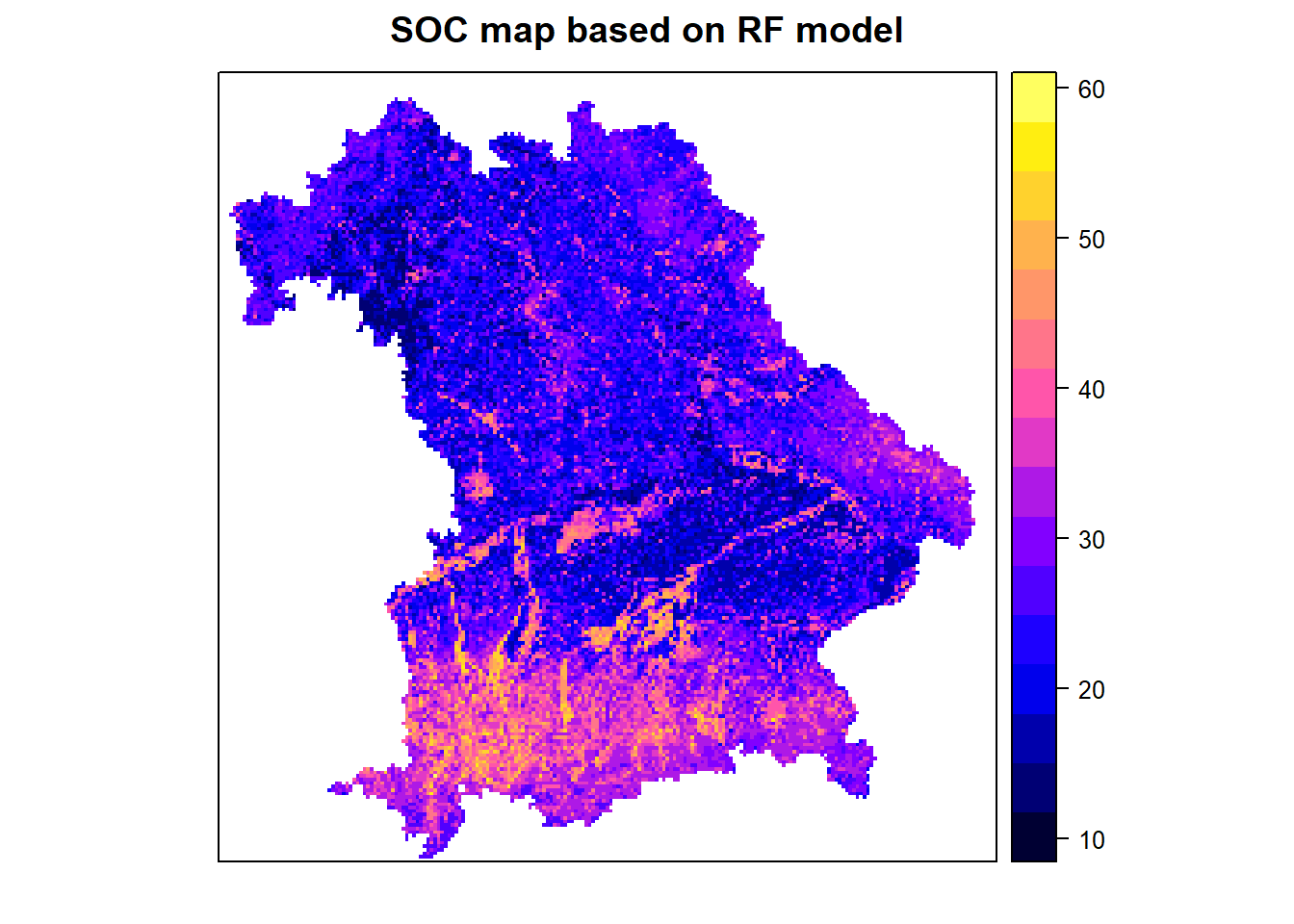
6.11 Tuning the models
A model can be improved in many ways, but if you cannot obtain better training data or covariates, tuning the model is always a good option. By tuning the model, you will choose the best parameters for your dataset. Careful, however, not to overfit your model specifically to your data, in which case it could not be reusable afterwards.
The caret package is often used because it allows the deployment of a variety of machine learning commands.
6.11.1 Setting the general training parameters
To have a better model, you can create a cross-validation. It will take a small set of your data every time and use it as a testing set while using the remaining part. To create a cross-validation, you need to set the trainControl(method = "cv", number = x) parameter before running your model. X will represent the number of folds you want to perform.
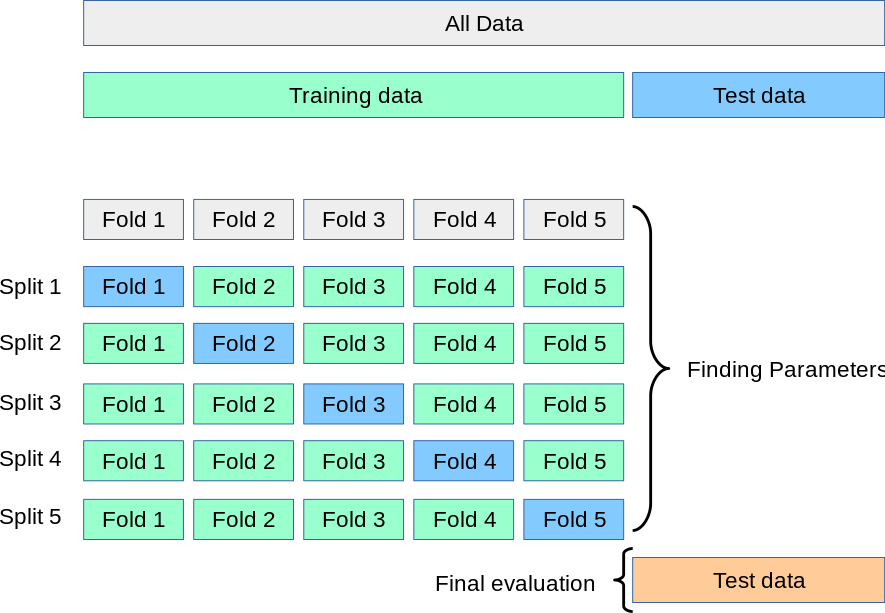
Figure 6.7: Cross validation fold schema.
Once the train control is set, you can create the training of the function with train(y ~ X, data =, method = "", trControl =, tuneGrid/tuneLength =, control =:
- y will be the column name of the data you want to predict, and X will be the predictors (you can replace it with a
.). The data is your training df. method = ""is the model you want to chose. You can look to allcaretpackage model on the webpage https://topepo.github.io/caret/available-models.html.trControl =correspond to thetrainControl()parameters we set earlier. -tuneGrid =is the tuning parameters of your model. You create a matrix of your tuning values withexpand.gridfunction. For each model, the tuning variables have specific names. You can also usetuneLength, which randomly creates a grid of the number you set root square, the number of tuning parameters for this model (not recommended). You can also set a sequence of several numbers in your grid.controlan additional command for extra control parameters.
#First control parameters
ctrl1 <- trainControl(method = "cv", number = 5)
set.seed(1234)
tree_fit1 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rpart",
trControl = ctrl1)
# display the results of decision tree
tree_fit1## CART
##
## 271 samples
## 12 predictor
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 215, 217, 217, 218, 217
## Resampling results across tuning parameters:
##
## cp RMSE Rsquared MAE
## 0.06162053 13.80990 0.19138312 10.31556
## 0.06856500 13.90378 0.18603785 10.39489
## 0.18912794 14.63942 0.09680234 11.41884
##
## RMSE was used to select the optimal model using the smallest value.
## The final value used for the model was cp = 0.06162053.## Decision tree model 1 had 5 Run in total#Second control parameters
ctrl2 <- trainControl(method = "cv", number = 20)
set.seed(1234)
tree_fit2 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rpart",
trControl = ctrl2)
# display the results of decision tree
tree_fit2## CART
##
## 271 samples
## 12 predictor
##
## No pre-processing
## Resampling: Cross-Validated (20 fold)
## Summary of sample sizes: 257, 256, 258, 258, 257, 256, ...
## Resampling results across tuning parameters:
##
## cp RMSE Rsquared MAE
## 0.06162053 13.77702 0.22658828 10.43208
## 0.06856500 14.07502 0.20270384 10.74035
## 0.18912794 14.85486 0.07527534 11.60622
##
## RMSE was used to select the optimal model using the smallest value.
## The final value used for the model was cp = 0.06162053.## Decision tree model 2 had 20 Run in totalYou can also repeat the cross-validation several time with the method = "repeatedcv"
#Third control parameters
ctrl3 <- trainControl(method = "repeatedcv", number = 5, repeats = 3)
set.seed(1234)
tree_fit3 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rpart",
trControl = ctrl3)
# display the results of decision tree
tree_fit3## CART
##
## 271 samples
## 12 predictor
##
## No pre-processing
## Resampling: Cross-Validated (5 fold, repeated 3 times)
## Summary of sample sizes: 215, 217, 217, 218, 217, 215, ...
## Resampling results across tuning parameters:
##
## cp RMSE Rsquared MAE
## 0.06162053 13.98031 0.18955436 10.46265
## 0.06856500 14.15751 0.17382230 10.58568
## 0.18912794 14.67725 0.09851222 11.38858
##
## RMSE was used to select the optimal model using the smallest value.
## The final value used for the model was cp = 0.06162053.## Decision tree model 3 had 15 Run in total6.11.2 Decision tree model tuning
rpart.control() Function allows you to tune your decision tree model:
cpis the complexity parameter that will set pruning. The lowest the number sits, the more complex the tree becomes.minsplitis the minimum number of observations needed for a split.minbucketis the minimum number of observations in a leaf.maxdepthis the maximum depth of the tree.
On the parameters of the decision tree, you can specify the tuneGrid parameter from the caret training with an expand.grid()
Figure 6.8: Decision tree parameters.
# Create a decision tree with simpler parameters
rpart_ctrl1 <- rpart.control(
cp = 0.05, # Complexity parameter
minsplit = 20, # Minimum number of observations to split
minbucket = 15, # Minimum number of observations in any terminal node
maxdepth = 4 # Maximum depth of the tree
)
set.seed(1234)
tree_fit4 <- rpart(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
method="anova", data=cov_soil_Train, control = rpart_ctrl1)
# display the results of the more simple decision tree
printcp(tree_fit4) ##
## Regression tree:
## rpart(formula = OC ~ CNBL + DEM + MCA + Slope + TWI + blue +
## green + red + nir + ndvi + rainfall + NPP, data = cov_soil_Train,
## method = "anova", control = rpart_ctrl1)
##
## Variables actually used in tree construction:
## [1] green NPP Slope
##
## Root node error: 61351/271 = 226.39
##
## n= 271
##
## CP nsplit rel error xerror xstd
## 1 0.189128 0 1.00000 1.01112 0.104709
## 2 0.068565 1 0.81087 0.84893 0.097943
## 3 0.061621 2 0.74231 0.79133 0.094026
## 4 0.050000 3 0.68069 0.78313 0.093608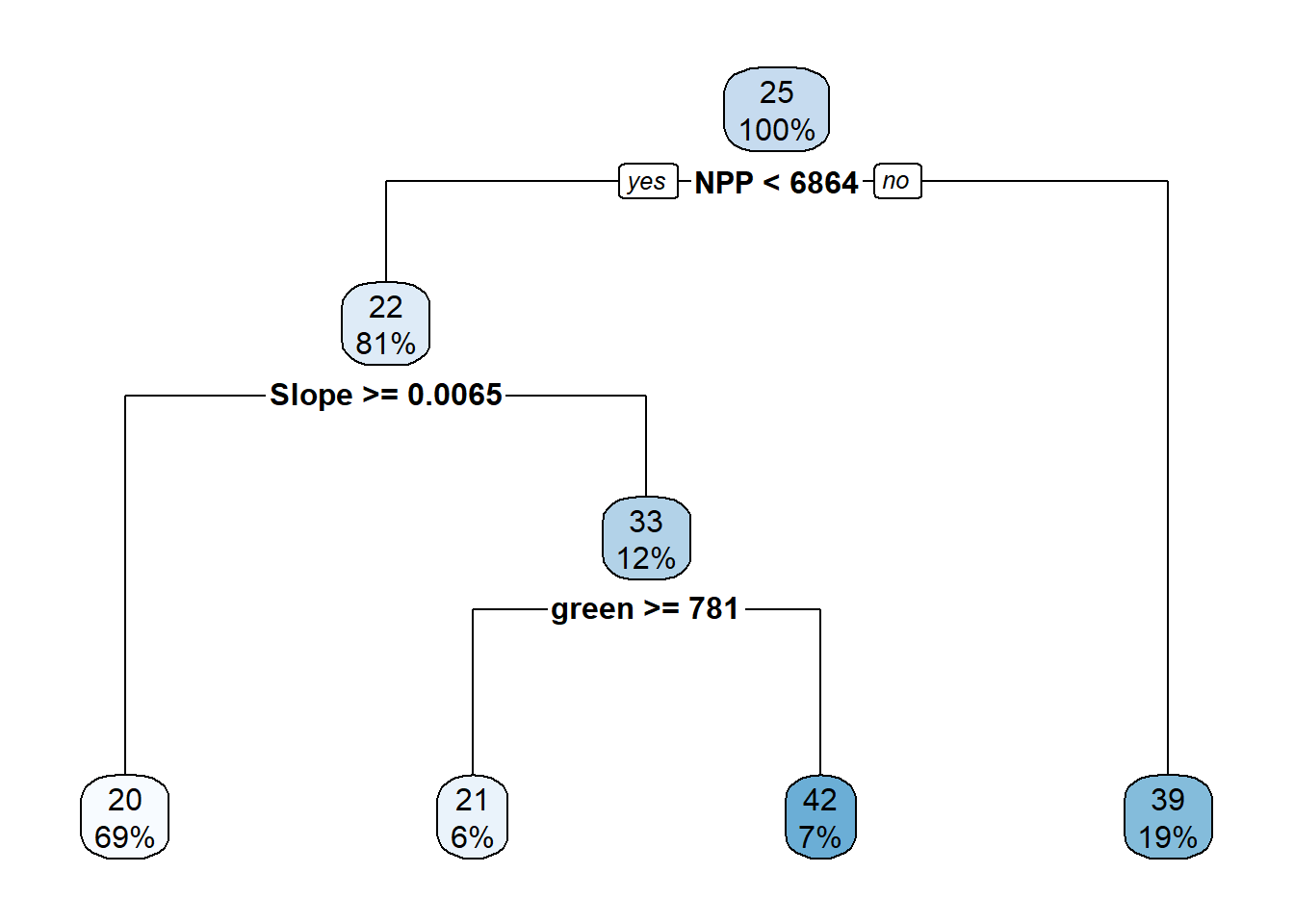
# Create a decision tree with more complex parameters
rpart_ctrl2 <- rpart.control(
cp = 0.0005, # Complexity parameter
minsplit = 10, # Minimum number of observations to split
minbucket = 5, # Minimum number of observations in any terminal node
maxdepth = 8 # Maximum depth of the tree
)
set.seed(1234)
tree_fit5 <- rpart(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
method="anova", data=cov_soil_Train, control = rpart_ctrl2)
# display the results of the more simple decision tree
printcp(tree_fit5) ##
## Regression tree:
## rpart(formula = OC ~ CNBL + DEM + MCA + Slope + TWI + blue +
## green + red + nir + ndvi + rainfall + NPP, data = cov_soil_Train,
## method = "anova", control = rpart_ctrl2)
##
## Variables actually used in tree construction:
## [1] blue CNBL DEM green MCA ndvi nir NPP
## [9] rainfall red Slope TWI
##
## Root node error: 61351/271 = 226.39
##
## n= 271
##
## CP nsplit rel error xerror xstd
## 1 0.1891279 0 1.00000 1.01112 0.104709
## 2 0.0685650 1 0.81087 0.84893 0.097943
## 3 0.0616205 2 0.74231 0.83187 0.096378
## 4 0.0434759 3 0.68069 0.80149 0.094160
## 5 0.0345742 4 0.63721 0.82250 0.095723
## 6 0.0333516 6 0.56806 0.81490 0.096209
## 7 0.0256923 7 0.53471 0.85861 0.103798
## 8 0.0221136 8 0.50902 0.85129 0.103001
## 9 0.0184032 9 0.48690 0.86388 0.102180
## 10 0.0163354 10 0.46850 0.83239 0.099363
## 11 0.0163285 11 0.45217 0.84990 0.100276
## 12 0.0151540 12 0.43584 0.84632 0.099720
## 13 0.0134087 13 0.42068 0.84857 0.100390
## 14 0.0118905 14 0.40727 0.83985 0.099958
## 15 0.0098702 15 0.39538 0.89144 0.109507
## 16 0.0087546 18 0.36577 0.92894 0.116296
## 17 0.0071396 19 0.35702 0.92501 0.116260
## 18 0.0063750 20 0.34988 0.93275 0.115394
## 19 0.0057885 21 0.34350 0.93703 0.115343
## 20 0.0046200 22 0.33772 0.95163 0.115323
## 21 0.0036255 23 0.33310 0.96827 0.116000
## 22 0.0031440 25 0.32584 0.96285 0.113771
## 23 0.0027501 26 0.32270 0.96343 0.113770
## 24 0.0019185 27 0.31995 0.96954 0.113830
## 25 0.0010823 28 0.31803 0.96873 0.113780
## 26 0.0009681 29 0.31695 0.97503 0.114070
## 27 0.0005000 30 0.31598 0.97487 0.114053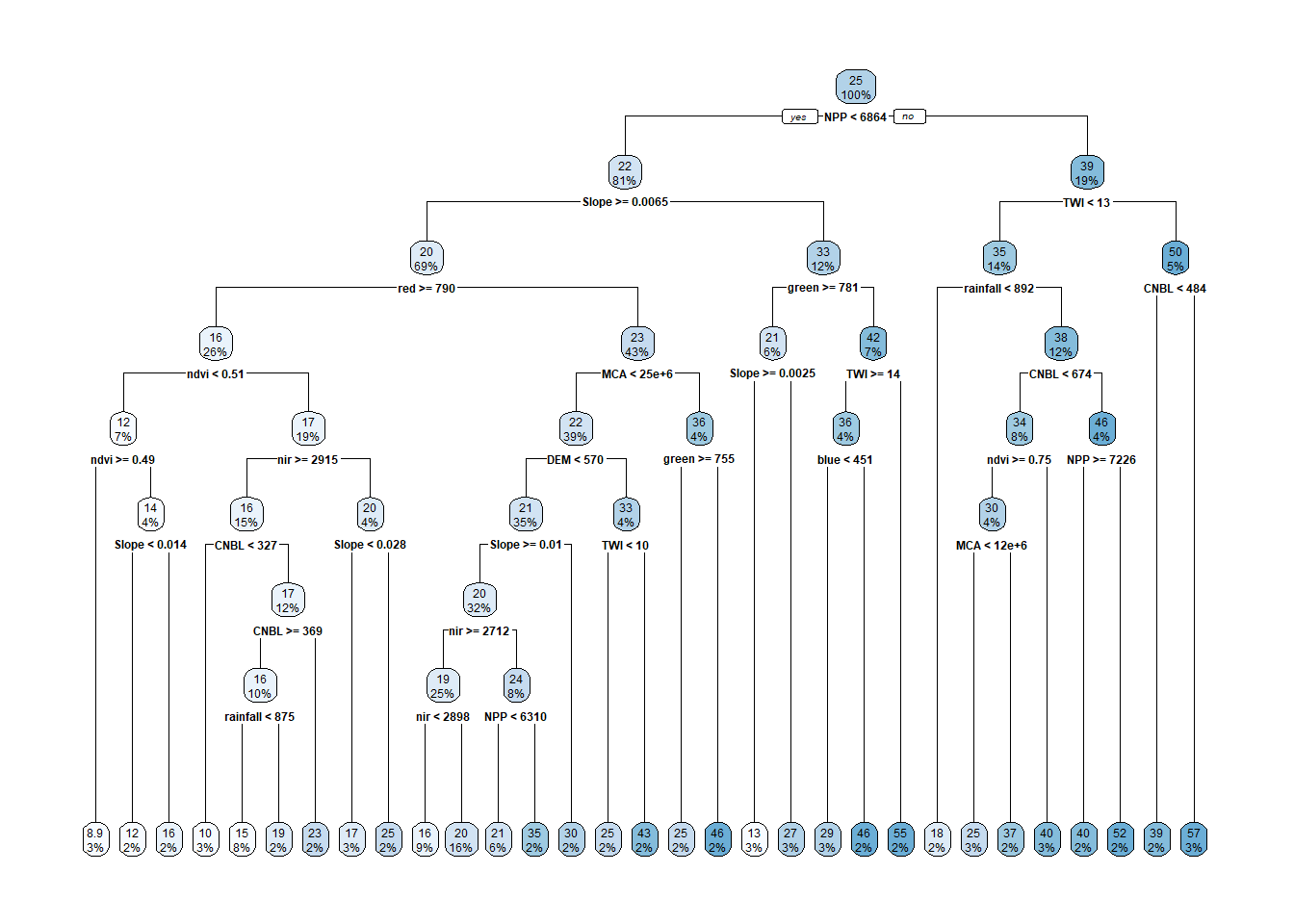
6.11.3 Random forest model tuning
For the random forest algorithm, you have two main tuning parameters:
ntreedecides the number of decision trees to generate. By default, it is set at 500, which is a good number, but you can change it with thentree =command line in the carettrainfunction.mtryis the number of variables selected for each split; the higher it is, the more complex your model becomes. In the carettrainfunction, you specify with thetrainGrid =parameter.
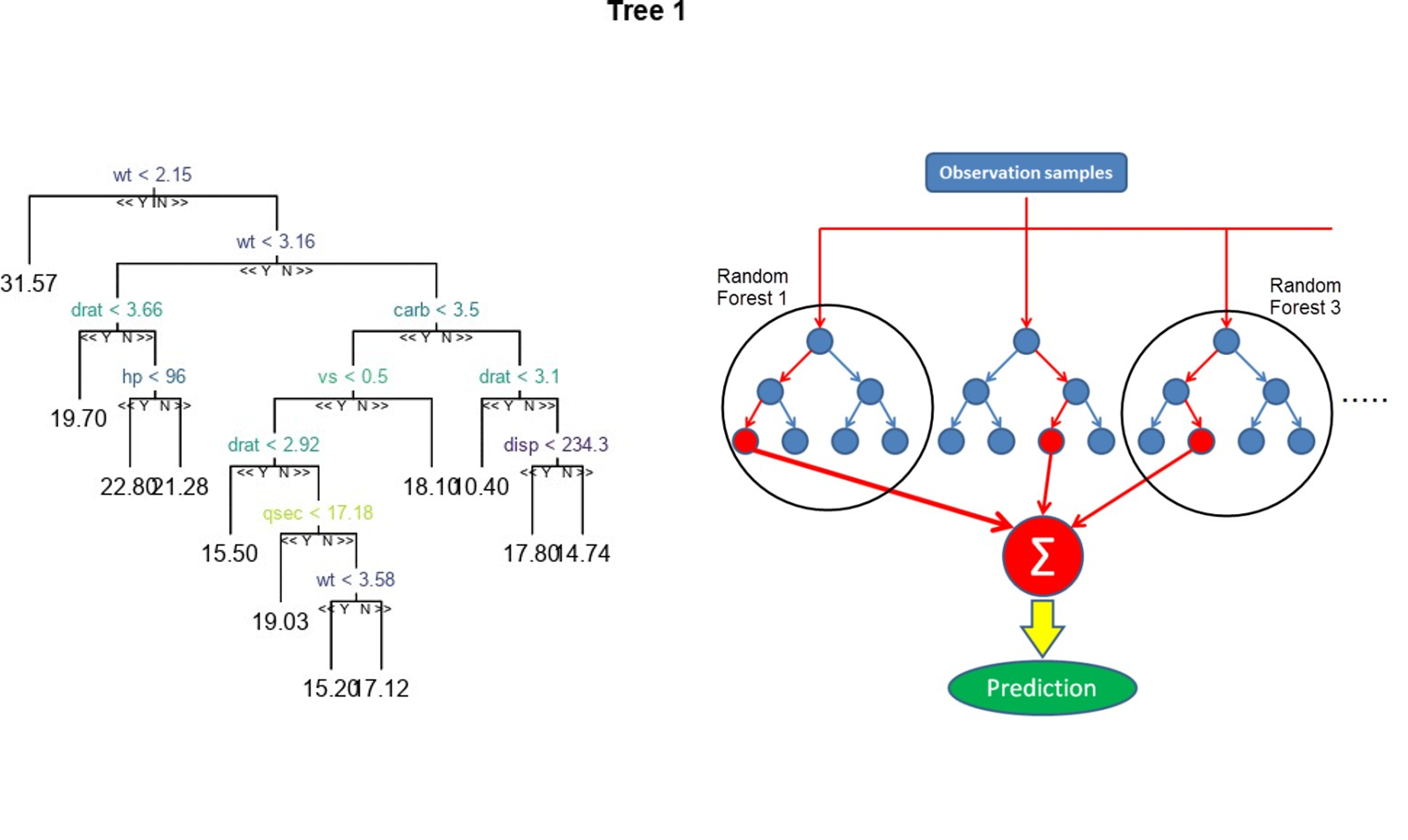
Figure 6.9: Random forest detailed schema.
# First RF tree with less complexity
ctrl <- trainControl(method = "cv", number = 10)
rfGrid1 <- expand.grid(.mtry = 3)
set.seed(1234)
rf_fit1 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rf",
trControl = ctrl,
tuneGrid = rfGrid1,
ntree = 200)
rf_fit1$finalModel##
## Call:
## randomForest(x = x, y = y, ntree = 200, mtry = param$mtry)
## Type of random forest: regression
## Number of trees: 200
## No. of variables tried at each split: 3
##
## Mean of squared residuals: 147.4249
## % Var explained: 34.88## mtry RMSE Rsquared MAE RMSESD RsquaredSD MAESD
## 1 3 11.67351 0.3984199 8.919224 2.01285 0.1343811 1.345439# Second RF tree with more complexity
rfGrid2 <- expand.grid(.mtry = 6)
set.seed(1234)
rf_fit2 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rf",
trControl = ctrl,
tuneGrid = rfGrid2,
ntree = 1000)
rf_fit2$finalModel##
## Call:
## randomForest(x = x, y = y, ntree = 1000, mtry = param$mtry)
## Type of random forest: regression
## Number of trees: 1000
## No. of variables tried at each split: 6
##
## Mean of squared residuals: 144.8437
## % Var explained: 36.02## mtry RMSE Rsquared MAE RMSESD RsquaredSD MAESD
## 1 6 11.72908 0.3922939 8.954656 1.889745 0.1179939 1.255245# Testing several tuning parameters at the same time
rfGrid3 <- expand.grid(.mtry = c(3:6))
set.seed(1234)
rf_fit3 <- train(OC ~ CNBL+DEM+MCA+Slope+TWI+blue+green+red+nir+ndvi+rainfall+NPP,
data = cov_soil_Train,
method = "rf",
trControl = ctrl,
tuneGrid = rfGrid3,
ntree = 500)
rf_fit3$finalModel##
## Call:
## randomForest(x = x, y = y, ntree = 500, mtry = param$mtry)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 4
##
## Mean of squared residuals: 143.2986
## % Var explained: 36.7## mtry RMSE Rsquared MAE RMSESD RsquaredSD MAESD
## 1 3 11.69037 0.3969708 8.892300 1.938197 0.1289380 1.268843
## 2 4 11.68114 0.3955169 8.896886 1.862187 0.1186351 1.237828
## 3 5 11.71240 0.3937666 8.912386 1.913175 0.1229028 1.264399
## 4 6 11.71687 0.3921174 8.908756 1.921134 0.1210158 1.2308576.12 Variables importance
There are several ways of assessing the variable importance of the model and the node impurity. The ** Gini** index or Gini ratio provides a ranking of the attributes for splitting the data. It stores the sum of the squared probabilities of each class or value. it can be used for classification problems.
\(Gini = 1 - \sum(P_i)^2\)
\(i\) = 1 to the number of classes. \(P\) = probabilities of each class.
With 0, the Gini impurity index is at its lowest. It occurs only when all the elements of the split are from the same class/value.
With a caret model trained you can use the varImp function to see the Gini index of all variables.
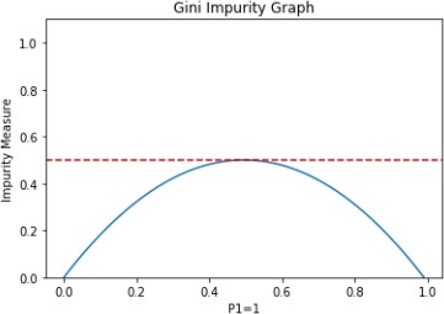
Figure 6.10: Gini impurity index.
\(\\[0.25cm]\)
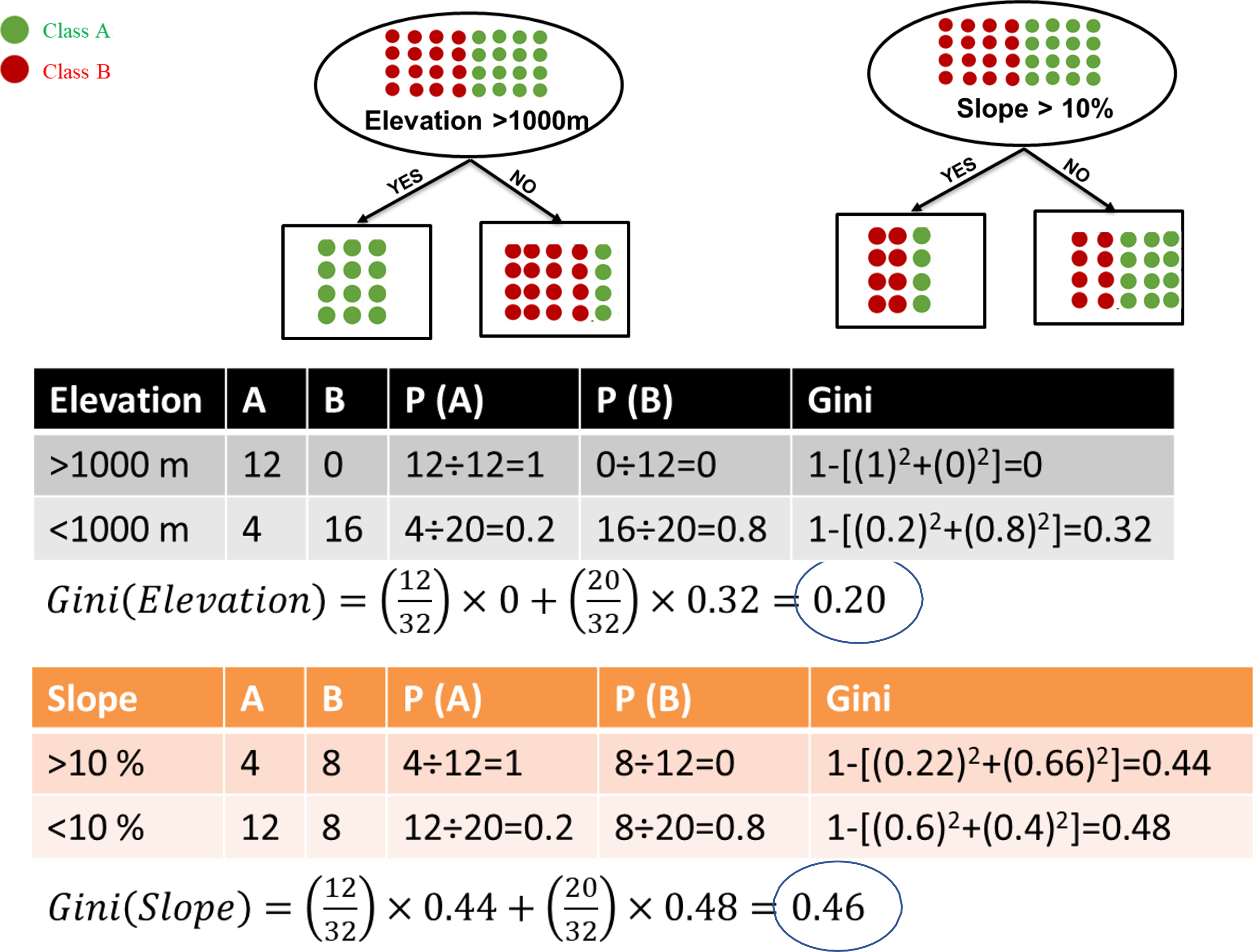
Figure 6.11: Gini formula example.
\(\\[0.2cm]\)
For regression problems MSE is a more adapted impurity measurement.
\(MSE = \frac{1}{n} \sum_{i=1}^{n}(Y_i - \hat{Y}_i)^2\)
\(n\) = the number of measurements. \(Y\) = the observed value. \(\hat{Y}\) = the predicted value
## rf variable importance
##
## Overall
## Slope 100.000
## TWI 91.693
## NPP 86.851
## MCA 78.089
## CNBL 75.723
## ndvi 73.076
## red 57.213
## nir 45.278
## green 42.216
## DEM 30.440
## blue 8.825
## rainfall 0.000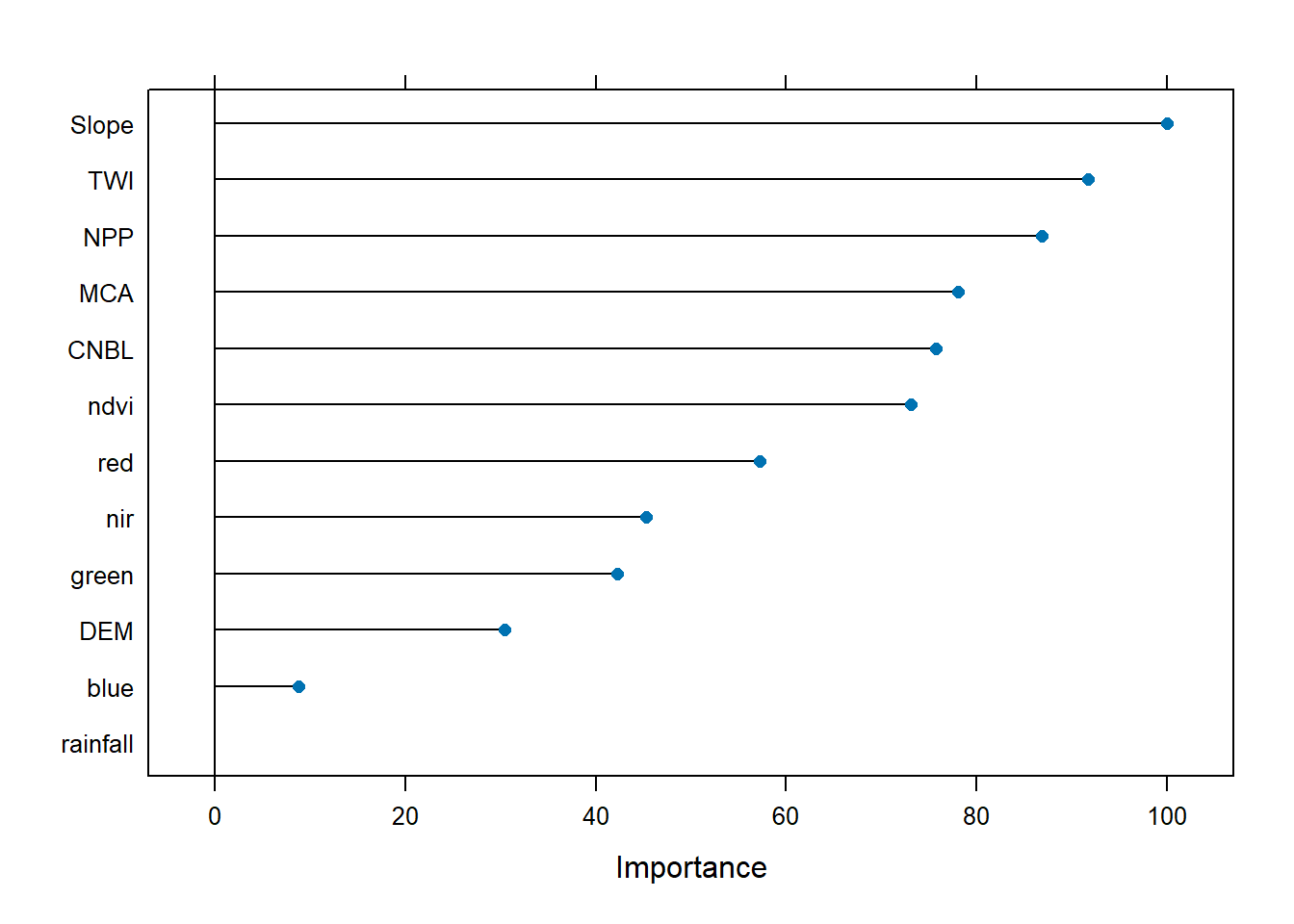
# More complex plot using ggplot function
# Set results in a df
importance_data <- as.data.frame(importance_rf$importance)
importance_data$Variable <- rownames(importance_data)
# Scaled it in percent and organising it in decreasing order
importance_data$Percentage <- importance_data$Overall / sum(importance_data$Overall) * 100
importance_data <- importance_data[order(importance_data$Percentage, decreasing = TRUE), ]
# Create the graph
importance_graph <- ggplot(importance_data, aes(x = reorder(Variable, Percentage), y = Percentage)) +
geom_bar(stat = "identity", fill = "skyblue", color = "black", width = 0.7, alpha = 0.7) +
coord_flip() +
labs(
title = "Importance of variables in percent",
x = "Variable",
y = "Importance (%)"
) +
theme_classic() +
theme(
axis.text = element_text(size = 12, color = "black"),
axis.title = element_text(size = 14, face = "bold"),
plot.title = element_text(size = 16, face = "bold", hjust = 0.5)
) +
geom_text(aes(label = round(Percentage, 1)), hjust = -0.2, color = "black", size = 4) +
ylim(0, max(importance_data$Percentage) * 1.1)
importance_graph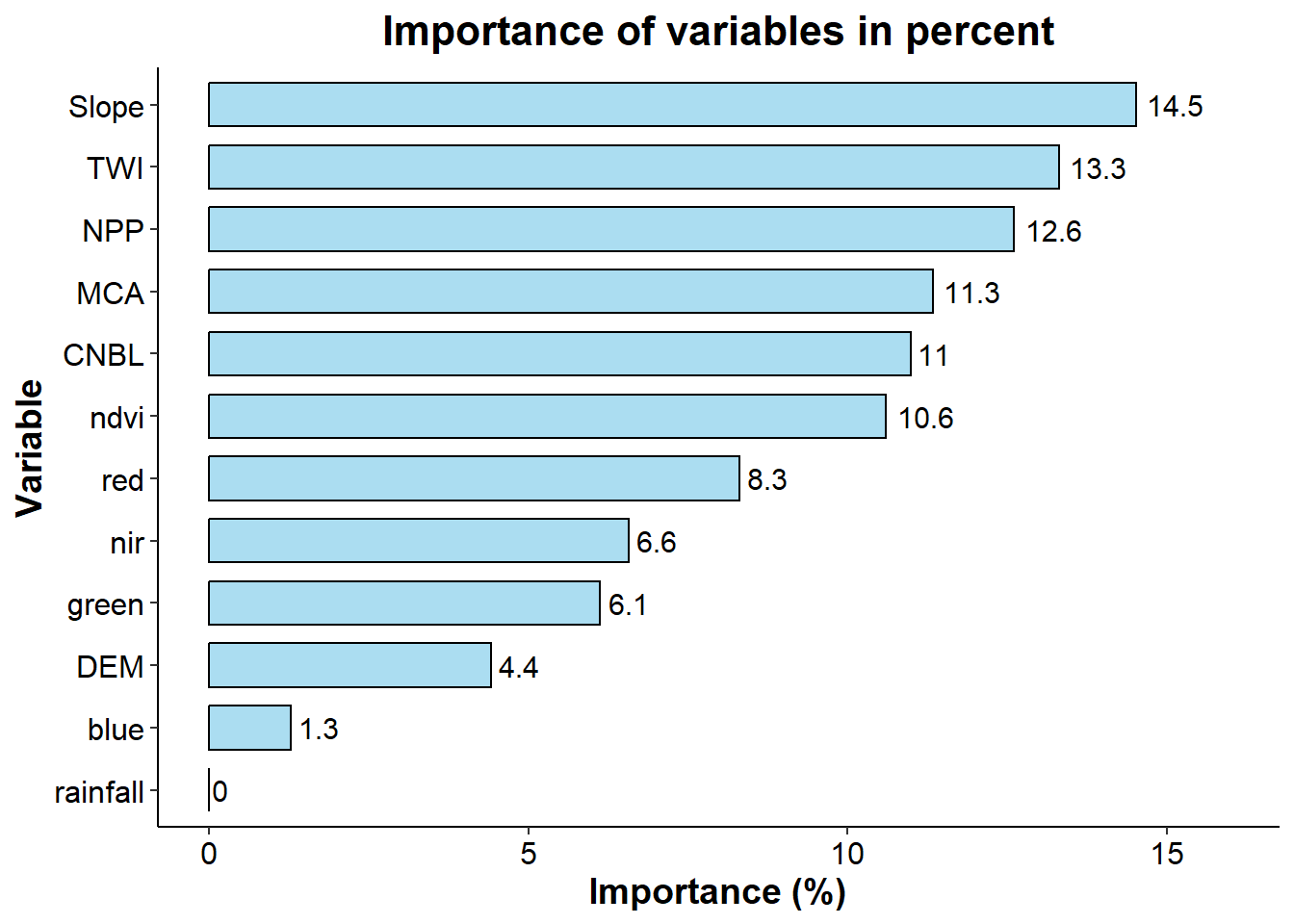
# Option to export the plot
#ggsave( "./Variables_importance_RF.pdf", importance_graph, width = 20, height = 13, units = "cm")